A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration
- PMID: 17884985
- PMCID: PMC1987388
- DOI: 10.1073/pnas.0703933104
A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration
Abstract
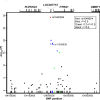
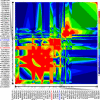
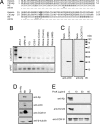
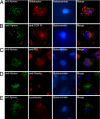
Genetic variants at chromosomes 1q31-32 and 10q26 are strongly associated with susceptibility to age-related macular degeneration (AMD), a common blinding disease of the elderly. We demonstrate, by evaluating 45 tag SNPs spanning HTRA1, PLEKHA1, and predicted gene LOC387715/ARMS2, that rs10490924 SNP alone, or a variant in strong linkage disequilibrium, can explain the bulk of association between the 10q26 chromosomal region and AMD. A previously suggested causal SNP, rs11200638, and other examined SNPs in the region are only indirectly associated with the disease. Contrary to previous reports, we show that rs11200638 SNP has no significant impact on HTRA1 promoter activity in three different cell lines, and HTRA1 mRNA expression exhibits no significant change between control and AMD retinas. However, SNP rs10490924 shows the strongest association with AMD (P = 5.3 x 10(-30)), revealing an estimated relative risk of 2.66 for GT heterozygotes and 7.05 for TT homozygotes. The rs10490924 SNP results in nonsynonymous A69S alteration in the predicted protein LOC387715/ARMS2, which has a highly conserved ortholog in chimpanzee, but not in other vertebrate sequences. We demonstrate that LOC387715/ARMS2 mRNA is detected in the human retina and various cell lines and encodes a 12-kDa protein, which localizes to the mitochondrial outer membrane when expressed in mammalian cells. We propose that rs10490924 represents a major susceptibility variant for AMD at 10q26. A likely biological mechanism is that the A69S change in the LOC387715/ARMS2 protein affects its presumptive function in mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Bringing the genetics of macular degeneration into focus.Proc Natl Acad Sci U S A. 2007 Oct 23;104(43):16725-6. doi: 10.1073/pnas.0708151104. Epub 2007 Oct 16. Proc Natl Acad Sci U S A. 2007. PMID: 17940012 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

