Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula
- PMID: 17885080
- PMCID: PMC2048810
- DOI: 10.1104/pp.107.107326
Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula
Abstract
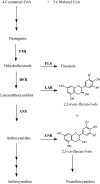
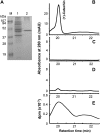
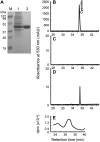
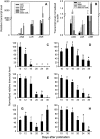
Oligomeric proanthocyanidins (PAs) composed primarily of epicatechin units accumulate in the seed coats of the model legume Medicago truncatula, reaching maximal levels at around 20 d after pollination. Genes encoding the single Medicago anthocyanidin synthase (ANS; EC 1.14.11.19) and leucoanthocyanidin reductase (LAR; EC 1.17.1.3) were cloned and the corresponding enzymes functionally identified. Recombinant MtANS converted leucocyanidin to cyanidin, and, more efficiently, dihydroquercetin to the flavonol quercetin. Levels of transcripts encoding dihydroflavonol reductase, ANS, and anthocyanidin reductase (ANR), the enzyme responsible for conversion of anthocyanidin to (-)-epicatechin, paralleled the accumulation of PAs in developing seeds, whereas LAR transcripts appeared to be more transiently expressed. LAR, ANS, and ANR proteins were localized to the cytosol in transfected tobacco (Nicotiana tabacum) leaves. Antisense down-regulation of ANS in M. truncatula resulted in reduced anthocyanin and PA levels, but had no impact on flavonol levels. Transgenic tobacco plants constitutively overexpressing MtLAR showed reduced anthocyanin content, but no catechin or increased levels of PAs were detected either in leaves or in flowers. Our results confirm previously ascribed in vivo functions for ANS and ANR. However, the apparent lack of catechin in M. truncatula PAs, the poor correlation between LAR expression and PA accumulation, and the lack of production of catechin monomers or oligomers in transgenic plants overexpressing MtLAR question the role of MtLAR in PA biosynthesis in Medicago.
Figures




Similar articles
-
The HD-ZIP IV transcription factor GLABRA2 acts as an activator for proanthocyanidin biosynthesis in Medicago truncatula seed coat.Plant J. 2024 Sep;119(5):2303-2315. doi: 10.1111/tpj.16918. Epub 2024 Jul 11. Plant J. 2024. PMID: 38990552
-
Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula.Plant Physiol. 2004 Mar;134(3):979-94. doi: 10.1104/pp.103.030221. Epub 2004 Feb 19. Plant Physiol. 2004. PMID: 14976232 Free PMC article.
-
Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase.BMC Plant Biol. 2013 Dec 5;13:202. doi: 10.1186/1471-2229-13-202. BMC Plant Biol. 2013. PMID: 24308601 Free PMC article.
-
Molecular Controls of Proanthocyanidin Synthesis and Structure: Prospects for Genetic Engineering in Crop Plants.J Agric Food Chem. 2018 Sep 26;66(38):9882-9888. doi: 10.1021/acs.jafc.8b02950. Epub 2018 Sep 13. J Agric Food Chem. 2018. PMID: 30139248 Review.
-
Revisiting decade-old questions in proanthocyanidin biosynthesis: current understanding and new challenges.Front Plant Sci. 2024 Mar 26;15:1373975. doi: 10.3389/fpls.2024.1373975. eCollection 2024. Front Plant Sci. 2024. PMID: 38595764 Free PMC article. Review.
Cited by
-
Metabolome and transcriptome profiling reveal regulatory network and mechanism of flavonoid biosynthesis during color formation of Dioscorea cirrhosa L.PeerJ. 2022 Jul 4;10:e13659. doi: 10.7717/peerj.13659. eCollection 2022. PeerJ. 2022. PMID: 35811818 Free PMC article.
-
Phenylpropanoid Content of Chickpea Seed Coats in Relation to Seed Dormancy.Plants (Basel). 2023 Jul 19;12(14):2687. doi: 10.3390/plants12142687. Plants (Basel). 2023. PMID: 37514301 Free PMC article.
-
Biosynthesis and genetic regulation of proanthocyanidins in plants.Molecules. 2008 Oct 28;13(10):2674-703. doi: 10.3390/molecules13102674. Molecules. 2008. PMID: 18971863 Free PMC article. Review.
-
A leucoanthocyanidin dioxygenase gene (RtLDOX2) from the feral forage plant Reaumuria trigyna promotes the accumulation of flavonoids and improves tolerance to abiotic stresses.J Plant Res. 2021 Sep;134(5):1121-1138. doi: 10.1007/s10265-021-01315-2. Epub 2021 May 26. J Plant Res. 2021. PMID: 34037878
-
The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway.Sci Rep. 2020 Aug 25;10(1):14161. doi: 10.1038/s41598-020-70977-0. Sci Rep. 2020. PMID: 32843672 Free PMC article.
References
-
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin P, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35 624–636 - PubMed
-
- Aerts RJ, Barry TN, McNabb WC (1999) Polyphenols and agriculture: beneficial effects of proanthocyanidins in forages. Agric Ecosyst Environ 75 1–12
-
- Antony U, Chandra TS (1998) Antinutrient reduction and enhancement in protein, starch, and mineral availability in fermented flour of finger millet (Eleusine coracana). J Agric Food Chem 46 2578–2582
-
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148 187–197 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

