Biosynthesis of phytosterol esters: identification of a sterol o-acyltransferase in Arabidopsis
- PMID: 17885082
- PMCID: PMC2048776
- DOI: 10.1104/pp.107.106278
Biosynthesis of phytosterol esters: identification of a sterol o-acyltransferase in Arabidopsis
Abstract
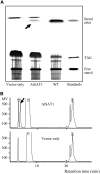
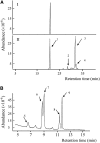
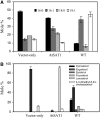
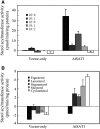
Fatty acyl esters of phytosterols are a major form of sterol conjugates distributed in many parts of plants. In this study we report an Arabidopsis (Arabidopsis thaliana) gene, AtSAT1 (At3g51970), which encodes for a novel sterol O-acyltransferase. When expressed in yeast (Saccharomyces cerevisiae), AtSAT1 mediated production of sterol esters enriched with lanosterol. Enzyme property assessment using cell-free lysate of yeast expressing AtSAT1 suggested the enzyme preferred cycloartenol as acyl acceptor and saturated fatty acyl-Coenyzme A as acyl donor. Taking a transgenic approach, we showed that Arabidopsis seeds overexpressing AtSAT1 accumulated fatty acyl esters of cycloartenol, accompanied by substantial decreases in ester content of campesterol and beta-sitosterol. Furthermore, fatty acid components of sterol esters from the transgenic lines were enriched with saturated and long-chain fatty acids. The enhanced AtSAT1 expression resulted in decreased level of free sterols, but the total sterol content in the transgenic seeds increased by up to 60% compared to that in wild type. We conclude that AtSAT1 mediates phytosterol ester biosynthesis, alternative to the route previously described for phospholipid:sterol acyltransferase, and provides the molecular basis for modification of phytosterol ester level in seeds.
Figures

Similar articles
-
Cellular sterol ester synthesis in plants is performed by an enzyme (phospholipid:sterol acyltransferase) different from the yeast and mammalian acyl-CoA:sterol acyltransferases.J Biol Chem. 2005 Oct 14;280(41):34626-34. doi: 10.1074/jbc.M504459200. Epub 2005 Jul 14. J Biol Chem. 2005. PMID: 16020547
-
Final report of the amended safety assessment of PEG-5, -10, -16, -25, -30, and -40 soy sterol.Int J Toxicol. 2004;23 Suppl 2:23-47. doi: 10.1080/10915810490499046. Int J Toxicol. 2004. PMID: 15513823 Review.
-
Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis.Plant Physiol. 2004 Jul;135(3):1324-35. doi: 10.1104/pp.104.044354. Epub 2004 Jul 9. Plant Physiol. 2004. PMID: 15247387 Free PMC article.
-
Aphid growth and reproduction on plants with altered sterol profiles: Novel insights using Arabidopsis mutant and overexpression lines.J Insect Physiol. 2020 May-Jun;123:104054. doi: 10.1016/j.jinsphys.2020.104054. Epub 2020 Apr 7. J Insect Physiol. 2020. PMID: 32275907
-
Biosynthesis and accumulation of sterols.Annu Rev Plant Biol. 2004;55:429-57. doi: 10.1146/annurev.arplant.55.031903.141616. Annu Rev Plant Biol. 2004. PMID: 15377227 Review.
Cited by
-
A distinct DGAT with sn-3 acetyltransferase activity that synthesizes unusual, reduced-viscosity oils in Euonymus and transgenic seeds.Proc Natl Acad Sci U S A. 2010 May 18;107(20):9464-9. doi: 10.1073/pnas.1001707107. Epub 2010 May 3. Proc Natl Acad Sci U S A. 2010. PMID: 20439724 Free PMC article.
-
Characterization of a Pentacyclic Triterpene Acetyltransferase Involved in the Biosynthesis of Taraxasterol and ψ-Taraxasterol Acetates in Lettuce.Front Plant Sci. 2022 Jan 3;12:788356. doi: 10.3389/fpls.2021.788356. eCollection 2021. Front Plant Sci. 2022. PMID: 35046976 Free PMC article.
-
Why Do Plants Convert Sitosterol to Stigmasterol?Front Plant Sci. 2019 Mar 28;10:354. doi: 10.3389/fpls.2019.00354. eCollection 2019. Front Plant Sci. 2019. PMID: 30984220 Free PMC article. Review.
-
Acyl-CoA-Binding Protein ACBP1 Modulates Sterol Synthesis during Embryogenesis.Plant Physiol. 2017 Jul;174(3):1420-1435. doi: 10.1104/pp.17.00412. Epub 2017 May 12. Plant Physiol. 2017. PMID: 28500265 Free PMC article.
-
Optimization of Campesterol-Producing Yeast Strains as a Feasible Platform for the Functional Reconstitution of Plant Membrane-Bound Enzymes.ACS Synth Biol. 2023 Apr 21;12(4):1109-1118. doi: 10.1021/acssynbio.2c00599. Epub 2023 Mar 27. ACS Synth Biol. 2023. PMID: 36972300 Free PMC article.
References
-
- Awad AB, Roy R, Fink CS (2003) β-sitosterol, a plant sterol, induces apoptosis and activates key caspases in MDA-MB-231 human breast cancer cells. Oncol Rep 10 497–500 - PubMed
-
- Awad AB, Smith AJ, Fink CS (2001) Plant sterols regulate rat vascular smooth muscle cell growth and prostacyclin release in culture. Prostaglandins Leukot Essent Fatty Acids 64 323–330 - PubMed
-
- Bach TJ (1986) Hydroxymethyl glutaryl-CoA reductase, a key enzyme in phytosterol synthesis? Lipids 21 82–88 - PubMed
-
- Bach TJ, Benveniste P (1997) Cloning of cDNAs or genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: heterologous expression and complementation analysis of mutations for functional characterization. Prog Lipid Res 36 197–226 - PubMed
-
- Banaś A, Carlsson AS, Huang B, Lenman M, Banas W, Lee M, Noiriel A, Benveniste P, Schaller H, Bouvier-Nave P, et al (2005) Cellular sterol ester synthesis in plants is performed by an enzyme (phospholipid:sterol acyltransferase) different from the yeast and mammalian acyl-CoA:sterol acyltransferases. J Biol Chem 280 34626–34634 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

