Unravelling the hidden heterogeneities of diffuse large B-cell lymphoma based on coupled two-way clustering
- PMID: 17888167
- PMCID: PMC2082044
- DOI: 10.1186/1471-2164-8-332
Unravelling the hidden heterogeneities of diffuse large B-cell lymphoma based on coupled two-way clustering
Abstract
Background: It becomes increasingly clear that our current taxonomy of clinical phenotypes is mixed with molecular heterogeneity. Of vital importance for refined clinical practice and improved intervention strategies is to define the hidden molecular distinct diseases using modern large-scale genomic approaches. Microarray omics technology has provided a powerful way to dissect hidden genetic heterogeneity of complex diseases. The aim of this study was thus to develop a bioinformatics approach to seek the transcriptional features leading to the hidden subtyping of a complex clinical phenotype. The basic strategy of the proposed method was to iteratively partition in two ways sample and feature space with super-paramagnetic clustering technique and to seek for hard and robust gene clusters that lead to a natural partition of disease samples and that have the highest functionally conceptual consensus evaluated with Gene Ontology.
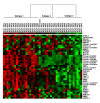
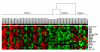
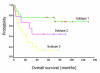
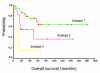
Results: We applied the proposed method to two publicly available microarray datasets of diffuse large B-cell lymphoma (DLBCL), a notoriously heterogeneous phenotype. A feature subset of 30 genes (38 probes) derived from analysis of the first dataset consisting of 4026 genes and 42 DLBCL samples identified three categories of patients with very different five-year overall survival rates (70.59%, 44.44% and 14.29% respectively; p = 0.0017). Analysis of the second dataset consisting of 7129 genes and 58 DLBCL samples revealed a feature subset of 13 genes (16 probes) that not only replicated the findings of the important DLBCL genes (e.g. JAW1 and BCL7A), but also identified three clinically similar subtypes (with 5-year overall survival rates of 63.13%, 34.92% and 15.38% respectively; p = 0.0009) to those identified in the first dataset. Finally, we built a multivariate Cox proportional-hazards prediction model for each feature subset and defined JAW1 as one of the most significant predictor (p = 0.005 and 0.014; hazard ratios = 0.02 and 0.03, respectively for two datasets) for both DLBCL cohorts under study.
Conclusion: Our results showed that the proposed algorithm is a promising computational strategy for peeling off the hidden genetic heterogeneity based on transcriptionally profiling disease samples, which may lead to an improved diagnosis and treatment of cancers.
Figures

References
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. - DOI - PubMed
-
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/S1535-6108(02)00032-6. - DOI - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI - PubMed
-
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

