The role of UbiX in Escherichia coli coenzyme Q biosynthesis
- PMID: 17889824
- PMCID: PMC2475804
- DOI: 10.1016/j.abb.2007.08.009
The role of UbiX in Escherichia coli coenzyme Q biosynthesis
Abstract
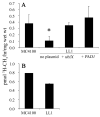
The reversible redox chemistry of coenzyme Q serves a crucial function in respiratory electron transport. Biosynthesis of Q in Escherichia coli depends on the ubi genes. However, very little is known about UbiX, an enzyme thought to be involved in the decarboxylation step in Q biosynthesis in E. coli and Salmonella enterica. Here we characterize an E. coli ubiX gene deletion strain, LL1, to further elucidate E. coli ubiX function in Q biosynthesis. LLI produces very low levels of Q, grows slowly on succinate as the sole carbon source, accumulates 4-hydroxy-3-octaprenyl-benzoate, and has reduced UbiG O-methyltransferase activity. Expression of either E. coli ubiX or the Saccharomyces cerevisiae ortholog PAD1, rescues the deficient phenotypes of LL1, identifying PAD1 as an ortholog of ubiX. Our results suggest that both UbiX and UbiD are required for the decarboxylation of 4-hydroxy-3-octaprenyl-benzoate in E. coli coenzyme Q biosynthesis, especially during logarithmic growth.
Figures
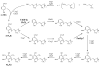





References
-
- Meganathan R. FEMS Microbiol Lett. 2001;203:131–139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

