Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation
- PMID: 17890163
- PMCID: PMC2692272
- DOI: 10.1016/j.mito.2007.08.001
Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation
Abstract
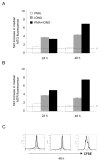
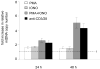
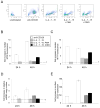
The quantity and activity of mitochondria vary dramatically in tissues and are modulated in response to changing cellular energy demands and environmental factors. The amount of mitochondrial DNA (mtDNA), which encodes essential subunits of the oxidative phosphorylation complexes required for cellular ATP production, is also tightly regulated, but by largely unknown mechanisms. Using murine T cells as a model system, we have addressed how specific signaling pathways influence mitochondrial biogenesis and mtDNA copy number. T cell receptor (TCR) activation results in a large increase in mitochondrial mass and membrane potential and a corresponding amplification of mtDNA, consistent with a vital role for mitochondrial function for growth and proliferation of these cells. Independent activation of protein kinase C (via PMA) or calcium-related pathways (via ionomycin) had differential and sub-maximal effects on these mitochondrial parameters, as did activation of naïve T cells with proliferative cytokines. Thus, the robust mitochondrial biogenesis response observed upon TCR activation requires synergy of multiple downstream signaling pathways. One such pathway involves AMP-activated protein kinase (AMPK), which we show has an unprecedented role in negatively regulating mitochondrial biogenesis that is mammalian target of rapamycin (mTOR)-dependent. That is, inhibition of AMPK after TCR signaling commences results in excessive, but uncoordinated mitochondrial proliferation. Thus mitochondrial biogenesis is not under control of a single master regulatory circuit, but rather requires the convergence of multiple signaling pathways with distinct downstream consequences on the organelle's structure, composition, and function.
Figures


References
-
- Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–93. - PubMed
-
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–97. - PubMed
-
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–25. - PubMed
-
- Burton TR, Kashour T, Wright JA, Amara FM. Cellular signaling pathways affect the function of ribonucleotide reductase mRNA binding proteins: mRNA stabilization, drug resistance, and malignancy (Review) Int J Oncol. 2003;22:21–31. - PubMed
-
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

