The dynamics of the roo transposable element in mutation-accumulation lines and segregating populations of Drosophila melanogaster
- PMID: 17890368
- PMCID: PMC2013678
- DOI: 10.1534/genetics.107.076174
The dynamics of the roo transposable element in mutation-accumulation lines and segregating populations of Drosophila melanogaster
Abstract
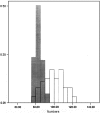
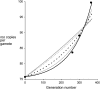
We estimated the number of copies for the long terminal repeat (LTR) retrotransposable element roo in a set of long-standing Drosophila melanogaster mutation-accumulation full-sib lines and in two large laboratory populations maintained with effective population size approximately 500, all of them derived from the same isogenic origin. Estimates were based on real-time quantitative PCR and in situ hybridization. Considering previous estimates of roo copy numbers obtained at earlier stages of the experiment, the results imply a strong acceleration of the insertion rate in the accumulation lines. The detected acceleration is consistent with a model where only one (maybe a few) of the approximately 70 roo copies in the ancestral isogenic genome was active and each active copy caused new insertions with a relatively high rate ( approximately 10(-2)), with new inserts being active copies themselves. In the two laboratory populations, however, a stabilized copy number or no accelerated insertion was found. Our estimate of the average deleterious viability effects per accumulated insert [E(s) < 0.003] is too small to account for the latter finding, and we discuss the mechanisms that could contain copy number.
Figures

References
-
- Abrusán, G., and H. J. Krambeck, 2006. Competition may determine the diversity of transposable elements. Theor. Popul. Biol. 70: 364–375. - PubMed
-
- Ávila, V., and A. García-Dorado, 2002. The effects of spontaneous mutation on competitive fitness in Drosophila melanogaster. J. Evol. Biol. 15: 561–566.
-
- Bartolomé, C., and X. Maside, 2004. The lack of recombination drives the fixation of transposable elements on fourth chromosome of Drosophila melanogaster. Genet. Res. 83: 91–100. - PubMed
-
- Biémont, C., and C. Vieira, 2006. Junk DNA as an evolutionary force. Nature 443: 521–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

