Downregulation of microRNAs-143 and -145 in B-cell malignancies
- PMID: 17892514
- PMCID: PMC11158757
- DOI: 10.1111/j.1349-7006.2007.00618.x
Downregulation of microRNAs-143 and -145 in B-cell malignancies
Abstract
Recently, it has been found that inappropriate expression of microRNAs (miRNAs) is strongly associated with carcinogenesis. In this study, we demonstrated that the expression of miRNAs (miRs) -143 and -145, the levels of which were previously shown to be reduced in colon cancers and various kinds of established cancer cell lines, was also decreased in most of the B-cell malignancies examined, including chronic lymphocytic leukemias (CLL), B-cell lymphomas, Epstein-Barr virus (EBV)-transformed B-cell lines, and Burkitt lymphoma cell lines. All samples from 13 CLL patients and eight of nine B-cell lymphoma ones tested exhibited an extremely low expression of miRs-143 and -145. The expression levels of miRs-143 and -145 were consistently low in human Burkitt lymphoma cell lines and were inversely associated with the cell proliferation observed in the EBV-transformed B-cell lines. Moreover, the introduction of either precursor or mature miR-143 and -145 into Raji cells resulted in a significant growth inhibition that occurred in a dose-dependent manner and the target gene of miRNA-143 was determined to be ERK5, as previously reported in human colon cancer DLD-1 cells. Taken together, these findings suggest that miRs-143 and -145 may be useful as biomarkers that differentiate B-cell malignant cells from normal cells and contribute to carcinogenesis in B-cell malignancies by a newly defined mechanism.
Figures

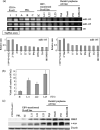

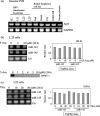
References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993; 75: 843–54. - PubMed
-
- Lagos‐Quintana M, Rauhut R, Lendeckel W, Tuschl T. Incorporating structure to predict microRNA targets. Science 2001; 294: 853–8. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001; 294: 858–62. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001; 294: 862–4. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

