Identifying HIV-1 dual infections
- PMID: 17892568
- PMCID: PMC2045676
- DOI: 10.1186/1742-4690-4-67
Identifying HIV-1 dual infections
Abstract
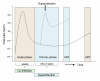
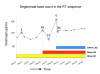
Transmission of human immunodeficiency virus (HIV) is no exception to the phenomenon that a second, productive infection with another strain of the same virus is feasible. Experiments with RNA viruses have suggested that both coinfections (simultaneous infection with two strains of a virus) and superinfections (second infection after a specific immune response to the first infecting strain has developed) can result in increased fitness of the viral population. Concerns about dual infections with HIV are increasing. First, the frequent detection of superinfections seems to indicate that it will be difficult to develop a prophylactic vaccine. Second, HIV-1 superinfections have been associated with accelerated disease progression, although this is not true for all persons. In fact, superinfections have even been detected in persons controlling their HIV infections without antiretroviral therapy. Third, dual infections can give rise to recombinant viruses, which are increasingly found in the HIV-1 epidemic. Recombinants could have increased fitness over the parental strains, as in vitro models suggest, and could exhibit increased pathogenicity. Multiple drug resistant (MDR) strains could recombine to produce a pan-resistant, transmittable virus. We will describe in this review what is presently known about super- and re-infection among ambient viral infections, as well as the first cases of HIV-1 superinfection, including HIV-1 triple infections. The clinical implications, the impact of the immune system, and the effect of anti-retroviral therapy will be covered, as will as the timing of HIV superinfection. The methods used to detect HIV-1 dual infections will be discussed in detail. To increase the likelihood of detecting a dual HIV-1 infection, pre-selection of patients can be done by serotyping, heteroduplex mobility assays (HMA), counting the degenerate base codes in the HIV-1 genotyping sequence, or surveying unexpected increases in the viral load during follow-up. The actual demonstration of dual infections involves a great deal of additional research to completely characterize the patient's viral quasispecies. The identification of a source partner would of course confirm the authenticity of the second infection.
Figures
References
-
- Roest RW, Carman WF, Maertzdorf J, Scoular A, Harvey J, Kant M, Van Der Meijden WI, Verjans GM, Osterhaus AD. Genotypic analysis of sequential genital herpes simplex virus type 1 (HSV-1) isolates of patients with recurrent HSV-1 associated genital herpes. J Med Virol. 2004;73:601–604. doi: 10.1002/jmv.20132. - DOI - PubMed
-
- Roest RW, Maertzdorf J, Kant M, Van Der Meijden WI, Osterhaus AD, Verjans GM. High incidence of genotypic variance between sequential herpes simplex virus type 2 isolates from HIV-1-seropositive patients with recurrent genital herpes. J Infect Dis. 2006;194:1115–1118. doi: 10.1086/507683. - DOI - PubMed
-
- Van Baarle D, Hovenkamp E, Kersten MJ, Klein MR, Miedema F, Van Oers MH. Direct Epstein-Barr virus (EBV) typing on peripheral blood mononuclear cells: no association between EBV type 2 infection or superinfection and the development of acquired immunodeficiency syndrome-related non-Hodgkin's lymphoma. Blood. 1999;93:3949–3955. - PubMed
-
- Drew WL, Sweet ES, Miner RC, Mocarski ES. Multiple infections by cytomegalovirus in patients with acquired immunodeficiency syndrome: documentation by Southern blot hybridization. J Infect Dis. 1984;150:952–953. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

