Cre reconstitution allows for DNA recombination selectively in dual-marker-expressing cells in transgenic mice
- PMID: 17893102
- PMCID: PMC2095822
- DOI: 10.1093/nar/gkm559
Cre reconstitution allows for DNA recombination selectively in dual-marker-expressing cells in transgenic mice
Abstract
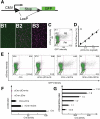
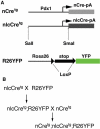
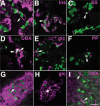
Cre/LoxP-based DNA recombination has been used to introduce desired DNA rearrangements in various organisms, having for example, greatly assisted genetic analyses in mice. For most applications, single gene promoters are used to drive Cre production for conditional gene activation/inactivation or lineage-tracing experiments. Such a manipulation introduces Cre in all cells in which the utilized promoter is active. To overcome the limited selectivity of single promoters for cell-type-specific recombination, we have explored the 'dual promoter combinatorial control' of Cre activity, so that Cre activity could be restricted to cells that express dual protein markers. We efficiently reconstituted Cre activity from two modified, inactive Cre fragments. Cre re-association was greatly enhanced by fusing the Cre fragments separately to peptides that can form a tight antiparallel leucine zipper. The co-expressed Cre fusion fragments showed substantial activity in cultured cells. As proof of principle of the utility of this technique in vivo for manipulating genes specifically in dual-marker-positive cells, we expressed each inactive Cre fragments in transgenic mice via individual promoters. Result showed the effective reconstitution of Cre activates LoxP recombination in the co-expressing cells.
Figures


Similar articles
-
Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast.Dev Dyn. 2002 Jun;224(2):245-51. doi: 10.1002/dvdy.10100. Dev Dyn. 2002. PMID: 12112477
-
Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation.Genesis. 2013 Jun;51(6):436-42. doi: 10.1002/dvg.22384. Epub 2013 Mar 26. Genesis. 2013. PMID: 23441020 Free PMC article.
-
Beta cell-specific ablation of target gene using Cre-loxP system in transgenic mice.J Surg Res. 1999 Jun 15;84(2):199-203. doi: 10.1006/jsre.1999.5642. J Surg Res. 1999. PMID: 10357920
-
Conditional mutagenesis in mice: the Cre/loxP recombination system.Int J Exp Pathol. 1996 Dec;77(6):269-78. Int J Exp Pathol. 1996. PMID: 9155661 Free PMC article. Review. No abstract available.
-
Pancreas-specific Cre driver lines and considerations for their prudent use.Cell Metab. 2013 Jul 2;18(1):9-20. doi: 10.1016/j.cmet.2013.06.011. Cell Metab. 2013. PMID: 23823474 Free PMC article. Review.
Cited by
-
Conditional transgenesis using Dimerizable Cre (DiCre).PLoS One. 2007 Dec 26;2(12):e1355. doi: 10.1371/journal.pone.0001355. PLoS One. 2007. PMID: 18159238 Free PMC article.
-
Split-Cre complementation restores combination activity on transgene excision in hair roots of transgenic tobacco.PLoS One. 2014 Oct 17;9(10):e110290. doi: 10.1371/journal.pone.0110290. eCollection 2014. PLoS One. 2014. PMID: 25329460 Free PMC article.
-
A step closer to making beta cells.Diabetologia. 2012 Mar;55(3):540-1. doi: 10.1007/s00125-011-2449-1. Epub 2012 Jan 13. Diabetologia. 2012. PMID: 22246379 No abstract available.
-
Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity.Dev Cell. 2019 Jan 7;48(1):49-63.e7. doi: 10.1016/j.devcel.2018.11.048. Dev Cell. 2019. PMID: 30620902 Free PMC article.
-
Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo.EMBO J. 2019 Jun 17;38(12):e102099. doi: 10.15252/embj.2019102099. Epub 2019 Apr 26. EMBO J. 2019. PMID: 31028085 Free PMC article.
References
-
- Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J. Biol. Chem. 1984;259:1509–1514. - PubMed
-
- Abremski K, Hoess R. Phage P1 Cre-loxP site-specific recombination. Effects of DNA supercoiling on catenation and knotting of recombinant products. J. Mol. Biol. 1985;184:211–220. - PubMed
-
- Gilbertson L. Cre-lox recombination: cre-ative tools for plant biotechnology. Trends Biotechnol. 2003;21:550–555. - PubMed
-
- Radhakrishnan P, Srivastava V. Utility of the FLP-FRT recombination system for genetic manipulation of rice. Plant Cell Rep. 2005;23:721–726. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

