Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design
- PMID: 17893128
- PMCID: PMC2168336
- DOI: 10.1128/IAI.00405-07
Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design
Abstract
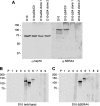
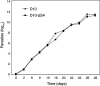
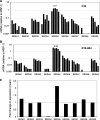
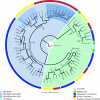
Serine repeat antigens (SERAs) are a family of secreted "cysteine-like" proteases of Plasmodium parasites. Several SERAs possess an atypical active-site serine residue in place of the canonical cysteine. The human malaria parasite Plasmodium falciparum possesses six "serine-type" (SERA1 to SERA5 and SERA9) and three "cysteine-type" (SERA6 to SERA8) SERAs. Here, we investigate the importance of the serine-type SERAs to blood-stage parasite development and examine the extent of functional redundancy among this group. We attempted to knock out the four P. falciparum serine-type SERA genes that have not been disrupted previously. SERA1, SERA4, and SERA9 knockout lines were generated, while only SERA5, the most strongly expressed member of the SERA family, remained refractory to genetic deletion. Interestingly, we discovered that while SERA4-null parasites completed the blood-stage cycle normally, they exhibited a twofold increase in the level of SERA5 mRNA. The inability to disrupt SERA5 and the apparent compensatory increase in SERA5 expression in response to the deletion of SERA4 provides evidence for an important blood-stage function for the serine-type SERAs and supports the notion of functional redundancy among this group. Such redundancy is consistent with our phylogenetic analysis, which reveals a monophyletic grouping of the serine-type SERAs across the genus Plasmodium and a predominance of postspeciation expansion. While SERA5 is to some extent further validated as a target for vaccine and drug development, our data suggest that the expression level of other serine-type SERAs is the only barrier to escape from anti-SERA5-specific interventions.
Figures


Similar articles
-
The Plasmodium falciparum pseudoprotease SERA5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes.PLoS Pathog. 2017 Jul 6;13(7):e1006453. doi: 10.1371/journal.ppat.1006453. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28683142 Free PMC article.
-
Enzymic, phylogenetic, and structural characterization of the unusual papain-like protease domain of Plasmodium falciparum SERA5.J Biol Chem. 2003 Nov 28;278(48):48169-77. doi: 10.1074/jbc.M306755200. Epub 2003 Sep 17. J Biol Chem. 2003. PMID: 13679369
-
A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle.J Biol Chem. 2002 Dec 6;277(49):47524-32. doi: 10.1074/jbc.M206974200. Epub 2002 Sep 11. J Biol Chem. 2002. PMID: 12228245
-
Characteristic features of the SERA multigene family in the malaria parasite.Parasit Vectors. 2020 Apr 6;13(1):170. doi: 10.1186/s13071-020-04044-y. Parasit Vectors. 2020. PMID: 32252804 Free PMC article. Review.
-
Plasmodium falciparum serine repeat antigen 5 (SE36) as a malaria vaccine candidate.Vaccine. 2011 Aug 11;29(35):5837-45. doi: 10.1016/j.vaccine.2011.06.052. Epub 2011 Jun 28. Vaccine. 2011. PMID: 21718740 Review.
Cited by
-
Protease-associated cellular networks in malaria parasite Plasmodium falciparum.BMC Genomics. 2011 Dec 23;12 Suppl 5(Suppl 5):S9. doi: 10.1186/1471-2164-12-S5-S9. Epub 2011 Dec 23. BMC Genomics. 2011. PMID: 22369208 Free PMC article.
-
The cellular and molecular basis for malaria parasite invasion of the human red blood cell.J Cell Biol. 2012 Sep 17;198(6):961-71. doi: 10.1083/jcb.201206112. J Cell Biol. 2012. PMID: 22986493 Free PMC article. Review.
-
Protease inhibitors from marine actinobacteria as a potential source for antimalarial compound.PLoS One. 2014 Mar 11;9(3):e90972. doi: 10.1371/journal.pone.0090972. eCollection 2014. PLoS One. 2014. PMID: 24618707 Free PMC article.
-
The Plasmodium falciparum pseudoprotease SERA5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes.PLoS Pathog. 2017 Jul 6;13(7):e1006453. doi: 10.1371/journal.ppat.1006453. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28683142 Free PMC article.
-
Molecular and biochemical characterization of a cathepsin B-like protease family unique to Trypanosoma congolense.Eukaryot Cell. 2008 Apr;7(4):684-97. doi: 10.1128/EC.00405-07. Epub 2008 Feb 15. Eukaryot Cell. 2008. PMID: 18281598 Free PMC article.
References
-
- Aoki, S., J. Li, S. Itagaki, B. A. Okech, T. G. Egwang, H. Matsuoka, N. M. Q. Palacpa, T. Mitamura, and T. Horii. 2002. Serine repeat antigen (SERA5) is predominantly expressed among the SERA multigene family of Plasmodium falciparum, and the acquired antibody titers correlate with serum inhibition of the parasite growth. J. Biol. Chem. 277:47533-47540. - PubMed
-
- Bourgon, R., M. Delorenzi, T. Sargeant, A. N. Hodder, B. S. Crabb, and T. P. Speed. 2004. The serine repeat antigen (SERA) gene family phylogeny in Plasmodium: the impact of GC content, and reconciliation of gene and species trees. Mol. Biol. Evol. 21:2161-2171. - PubMed
-
- Cowman, A. F., and B. S. Crabb. 2006. Invasion of red blood cells by malaria parasites. Cell 124:755-766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

