Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly
- PMID: 17893323
- PMCID: PMC2169193
- DOI: 10.1128/MCB.01213-07
Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly
Abstract
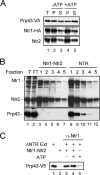
The Saccharomyces cerevisiae splicing factors Ntr1 (also known as Spp382) and Ntr2 form a stable complex and can further associate with DExD/H-box RNA helicase Prp43 to form a functional complex, termed the NTR complex, which catalyzes spliceosome disassembly. We show that Prp43 interacts with Ntr1-Ntr2 in a dynamic manner. The Ntr1-Ntr2 complex can also bind to the spliceosome first, before recruiting Prp43 to catalyze disassembly. Binding of Ntr1-Ntr2 or Prp43 does not require ATP, but disassembly of the spliceosome requires hydrolysis of ATP. The NTR complex also dynamically interacts with U5 snRNP. Ntr2 interacts with U5 component Brr2 and is essential for both interactions of NTR with U5 and with the spliceosome. Ntr2 alone can also bind to U5 and to the spliceosome, suggesting a role of Ntr2 in mediating the binding of NTR to the spliceosome through its interaction with U5. Our results demonstrate that dynamic interactions of NTR with U5, through the interaction of Ntr2 with Brr2, and interactions of Ntr1 and Prp43 govern the recruitment of Prp43 to the spliceosome to mediate spliceosome disassembly.
Figures




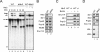


References
-
- Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. - PubMed
-
- Burge, C. B., T. H. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Burgess, S. M., and C. Guthrie. 1993. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73:1377-1392. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
