ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription
- PMID: 17893325
- PMCID: PMC2169170
- DOI: 10.1128/MCB.00972-07
ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription
Erratum in
- Mol Cell Biol. 2009 May;29(9):2481
Abstract
Cytoplasmic mRNA localization regulates gene expression by spatially restricting protein translation. Recent evidence has shown that nuclear proteins (such as hnRNPs) are required to form mRNPs capable of cytoplasmic localization. ZBP1 and ZBP2, two hnRNP K homology domain-containing proteins, were previously identified by their binding to the zipcode, the sequence element necessary and sufficient for beta-actin mRNA localization. ZBP1 colocalizes with nascent beta-actin mRNA in the nucleus but is predominantly a cytoplasmic protein. ZBP2, in contrast, is predominantly nuclear. We hypothesized that the two proteins cooperate to localize beta-actin mRNA and sought to address where and how this might occur. We demonstrate that ZBP2, a homologue of the splicing factor KSRP, binds initially to nascent beta-actin transcripts and facilitates the subsequent binding of the shuttling ZBP1. ZBP1 then associates with the RNA throughout the nuclear export and cytoplasmic localization process.
Figures






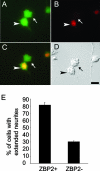
Similar articles
-
A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons.J Cell Biol. 2002 Jan 7;156(1):41-51. doi: 10.1083/jcb.200105133. Epub 2002 Jan 7. J Cell Biol. 2002. PMID: 11781334 Free PMC article.
-
Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1.Nature. 2005 Nov 24;438(7067):512-5. doi: 10.1038/nature04115. Nature. 2005. PMID: 16306994
-
Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment.J Cell Biol. 2003 Jan 6;160(1):77-87. doi: 10.1083/jcb.200206003. Epub 2002 Dec 30. J Cell Biol. 2003. PMID: 12507992 Free PMC article.
-
ELAV proteins along evolution: back to the nucleus?Mol Cell Neurosci. 2013 Sep;56:447-55. doi: 10.1016/j.mcn.2013.02.003. Epub 2013 Feb 22. Mol Cell Neurosci. 2013. PMID: 23439364 Review.
-
A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts.Mol Cell Biol. 2005 Jul;25(13):5307-16. doi: 10.1128/MCB.25.13.5307-5316.2005. Mol Cell Biol. 2005. PMID: 15964789 Free PMC article. Review. No abstract available.
Cited by
-
mRNA localization: an orchestration of assembly, traffic and synthesis.Traffic. 2013 Jan;14(1):2-14. doi: 10.1111/tra.12004. Epub 2012 Sep 13. Traffic. 2013. PMID: 22913533 Free PMC article. Review.
-
The functional organization of axonal mRNA transport and translation.Nat Rev Neurosci. 2021 Feb;22(2):77-91. doi: 10.1038/s41583-020-00407-7. Epub 2020 Dec 7. Nat Rev Neurosci. 2021. PMID: 33288912 Free PMC article. Review.
-
NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure.Elife. 2013 Jan 22;2:e00178. doi: 10.7554/eLife.00178. Elife. 2013. PMID: 23359859 Free PMC article.
-
Molecular mechanisms behind mRNA localization in axons.Open Biol. 2020 Sep;10(9):200177. doi: 10.1098/rsob.200177. Epub 2020 Sep 23. Open Biol. 2020. PMID: 32961072 Free PMC article. Review.
-
KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons.PLoS One. 2013 Nov 14;8(11):e79255. doi: 10.1371/journal.pone.0079255. eCollection 2013. PLoS One. 2013. PMID: 24244461 Free PMC article.
References
-
- Adams, M. D., R. S. Tarng, and D. C. Rio. 1997. The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes. Dev. 11:129-138. - PubMed
-
- Bassell, G. J., and R. H. Singer. 2001. Neuronal RNA localization and the cytoskeleton. Probl. Cell Differ. 34:41-56. - PubMed
-
- Carey, J., V. Cameron, P. L. de Haseth, and O. C. Uhlenbeck. 1983. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry 22:2601-2610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
