Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions
- PMID: 17893328
- PMCID: PMC2169176
- DOI: 10.1128/MCB.00899-07
Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions
Abstract
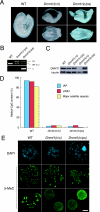
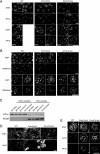
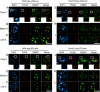
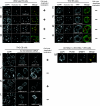
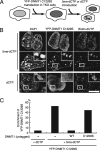
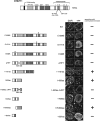
DNA methyltransferase 1 (DNMT1) plays an important role in the inheritance of genomic DNA methylation, which is coupled to the DNA replication process. Early embryonic lethality in DNMT1-null mutant (Dnmt1(c)) mice indicates that DNA methylation is essential for mammalian development. DNMT1, however, interacts with a number of transcriptional regulators and has a transcriptional repressor activity independent of its catalytic activity. To examine the roles of the catalytic activity of DNMT1 in vivo, we generated a Dnmt1(ps) allele that expresses a point-mutated protein that lacks catalytic activity (DNMT1-C1229S). Dnmt1(ps) mutant mice showed developmental arrest shortly after gastrulation, near-complete loss of DNA methylation, and an altered distribution of repressive chromatin markers in the nuclei; these phenotypes are quite similar to those of the Dnmt1(c) mutant. The mutant DNMT1 protein failed to associate with replication foci in Dnmt1(ps) cells. Reconstitution experiments and replication labeling in Dnmt1-/- Dnmt3a-/- Dnmt3b-/- (i.e., unmethylated) embryonic stem cells revealed that preexisting DNA methylation is a major determinant for the cell cycle-dependent localization of DNMT1. The C-terminal catalytic domain of DNMT1 inhibited its stable association with unmethylated chromatin. Our results reveal essential roles for the DNA methylation mark in mammalian development and in DNMT1 localization.
Figures



References
-
- Araujo, F. D., S. Croteau, A. D. Slack, S. Milutinovic, P. Bigey, G. B. Price, M. Zannis-Hajopoulos, and M. Szyf. 2001. The DNMT1 target recognition domain resides in the N terminus. J. Biol. Chem. 276:6930-6936. - PubMed
-
- Bacolla, A., S. Pradhan, R. J. Roberts, and R. D. Wells. 1999. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated dna. J. Biol. Chem. 274:33011-33019. - PubMed
-
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. - PubMed
-
- Chen, T., S. Hevi, F. Gay, N. Tsujimoto, T. He, B. Zhang, Y. Ueda, and E. Li. 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39:391-396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
