Reduced C(beta) statistical potentials can outperform all-atom potentials in decoy identification
- PMID: 17893359
- PMCID: PMC2204143
- DOI: 10.1110/ps.072939707
Reduced C(beta) statistical potentials can outperform all-atom potentials in decoy identification
Abstract
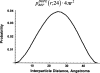
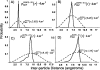
We developed a series of statistical potentials to recognize the native protein from decoys, particularly when using only a reduced representation in which each side chain is treated as a single C(beta) atom. Beginning with a highly successful all-atom statistical potential, the Discrete Optimized Protein Energy function (DOPE), we considered the implications of including additional information in the all-atom statistical potential and subsequently reducing to the C(beta) representation. One of the potentials includes interaction energies conditional on backbone geometries. A second potential separates sequence local from sequence nonlocal interactions and introduces a novel reference state for the sequence local interactions. The resultant potentials perform better than the original DOPE statistical potential in decoy identification. Moreover, even upon passing to a reduced C(beta) representation, these statistical potentials outscore the original (all-atom) DOPE potential in identifying native states for sets of decoys. Interestingly, the backbone-dependent statistical potential is shown to retain nearly all of the information content of the all-atom representation in the C(beta) representation. In addition, these new statistical potentials are combined with existing potentials to model hydrogen bonding, torsion energies, and solvation energies to produce even better performing potentials. The ability of the C(beta) statistical potentials to accurately represent protein interactions bodes well for computational efficiency in protein folding calculations using reduced backbone representations, while the extensions to DOPE illustrate general principles for improving knowledge-based potentials.
Figures



Similar articles
-
Improving the orientation-dependent statistical potential using a reference state.Proteins. 2014 Oct;82(10):2383-93. doi: 10.1002/prot.24600. Epub 2014 Jun 3. Proteins. 2014. PMID: 24810843
-
An accurate, residue-level, pair potential of mean force for folding and binding based on the distance-scaled, ideal-gas reference state.Protein Sci. 2004 Feb;13(2):400-11. doi: 10.1110/ps.03348304. Protein Sci. 2004. PMID: 14739325 Free PMC article.
-
Protein refolding in silico with atom-based statistical potentials and conformational search using a simple genetic algorithm.J Mol Biol. 2006 Jun 23;359(5):1456-67. doi: 10.1016/j.jmb.2006.04.033. Epub 2006 Apr 27. J Mol Biol. 2006. PMID: 16678202
-
Statistical potential for assessment and prediction of protein structures.Protein Sci. 2006 Nov;15(11):2507-24. doi: 10.1110/ps.062416606. Protein Sci. 2006. PMID: 17075131 Free PMC article.
-
Protein modeling with reduced representation: statistical potentials and protein folding mechanism.Acta Biochim Pol. 2005;52(4):741-8. Epub 2005 May 31. Acta Biochim Pol. 2005. PMID: 15933762 Review.
Cited by
-
How well can the accuracy of comparative protein structure models be predicted?Protein Sci. 2008 Nov;17(11):1881-93. doi: 10.1110/ps.036061.108. Epub 2008 Oct 1. Protein Sci. 2008. PMID: 18832340 Free PMC article.
-
Explicit orientation dependence in empirical potentials and its significance to side-chain modeling.Acc Chem Res. 2009 Aug 18;42(8):1087-96. doi: 10.1021/ar900009e. Acc Chem Res. 2009. PMID: 19445451 Free PMC article.
-
A probabilistic and continuous model of protein conformational space for template-free modeling.J Comput Biol. 2010 Jun;17(6):783-98. doi: 10.1089/cmb.2009.0235. J Comput Biol. 2010. PMID: 20583926 Free PMC article.
-
Automated real-space refinement of protein structures using a realistic backbone move set.Biophys J. 2011 Aug 17;101(4):899-909. doi: 10.1016/j.bpj.2011.06.063. Biophys J. 2011. PMID: 21843481 Free PMC article.
-
On docking, scoring and assessing protein-DNA complexes in a rigid-body framework.PLoS One. 2012;7(2):e32647. doi: 10.1371/journal.pone.0032647. Epub 2012 Feb 29. PLoS One. 2012. PMID: 22393431 Free PMC article.
References
-
- Anfinsen C.B. 1973. Principles that govern folding of protein chains. Science 181: 223–230. - PubMed
-
- Bauer A. and Beyer, A. 1994. An improved pair potential to recognize native protein folds. Proteins 18: 254–261. - PubMed
-
- Bennaim A. 1997. Statistical potentials extracted from protein structures: Are these meaningful potentials? J. Chem. Phys. 107: 3698–3706.
-
- Betancourt M.R. 2003. A reduced protein model with accurate native-structure identification ability. Proteins 53: 889–907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

