Forbidden penta-peptides
- PMID: 17893362
- PMCID: PMC2204130
- DOI: 10.1110/ps.073067607
Forbidden penta-peptides
Abstract
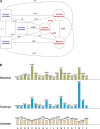
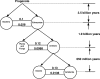
There are 3,200,000 amino acid sequences of length 5 (penta-peptides). Statistically, we expect to see a distribution of penta-peptides that is determined by the frequency of the participating amino acids. We show, however, that not only are there thousands of such penta-peptides that are absent from all known proteomes, but many of them are coded for multiple times in the non-coding genomic regions. This suggests a strong selection process that prevents these peptides from being expressed. We also show that the characteristics of these forbidden penta-peptides vary among different phylogenetic groups (e.g., eukaryotes, prokaryotes, and archaea). Our analysis provides the first steps toward understanding the "grammar" of the forbidden penta-peptides.
Figures

References
-
- Abe N. and Mamitsuka, H. 1997. Predicting protein secondary structure using stochastic tree grammars. J Mach Learn 29: 275–301.
-
- Alberts B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. 2002. Molecular biology of the cell, 4th ed. Garland, New York.
-
- Benjamini Y. and Yekutieli, D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 29: 1165–1188.
-
- Blaber M., Zhang, X.J., and Matthews, B.W. 1993. Structural basis of amino acid alpha helix propensity. Science 260: 1637–1640. - PubMed
-
- Bystroff C., Thorsson, V., and Baker, D. 2000. HMMSTR: A hidden Markov model for local sequence-structure correlations in proteins. J. Mol. Biol. 301: 173–190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

