Interspecies hybridization on DNA resequencing microarrays: efficiency of sequence recovery and accuracy of SNP detection in human, ape, and codfish mitochondrial DNA genomes sequenced on a human-specific MitoChip
- PMID: 17894875
- PMCID: PMC2211321
- DOI: 10.1186/1471-2164-8-339
Interspecies hybridization on DNA resequencing microarrays: efficiency of sequence recovery and accuracy of SNP detection in human, ape, and codfish mitochondrial DNA genomes sequenced on a human-specific MitoChip
Abstract
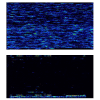
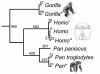
Background: Iterative DNA "resequencing" on oligonucleotide microarrays offers a high-throughput method to measure intraspecific iodiversity, one that is especially suited to SNP-dense gene regions such as vertebrate mitochondrial (mtDNA) genomes. However, costs of single-species design and microarray fabrication are prohibitive. A cost-effective, multi-species strategy is to hybridize experimental DNAs from diverse species to a common microarray that is tiled with oligonucleotide sets from multiple, homologous reference genomes. Such a strategy requires that cross-hybridization between the experimental DNAs and reference oligos from the different species not interfere with the accurate recovery of species-specific data. To determine the pattern and limits of such interspecific hybridization, we compared the efficiency of sequence recovery and accuracy of SNP identification by a 15,452-base human-specific microarray challenged with human, chimpanzee, gorilla, and codfish mtDNA genomes.
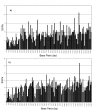
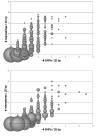
Results: In the human genome, 99.67% of the sequence was recovered with 100.0% accuracy. Accuracy of SNP identification declines log-linearly with sequence divergence from the reference, from 0.067 to 0.247 errors per SNP in the chimpanzee and gorilla genomes, respectively. Efficiency of sequence recovery declines with the increase of the number of interspecific SNPs in the 25b interval tiled by the reference oligonucleotides. In the gorilla genome, which differs from the human reference by 10%, and in which 46% of these 25b regions contain 3 or more SNP differences from the reference, only 88% of the sequence is recoverable. In the codfish genome, which differs from the reference by > 30%, less than 4% of the sequence is recoverable, in short islands > or = 12b that are conserved between primates and fish.
Conclusion: Experimental DNAs bind inefficiently to homologous reference oligonucleotide sets on a re-sequencing microarray when their sequences differ by more than a few percent. The data suggest that interspecific cross-hybridization will not interfere with the accurate recovery of species-specific data from multispecies microarrays, provided that the species' DNA sequences differ by > 20% (mean of 5b differences per 25b oligo). Recovery of DNA sequence data from multiple, distantly-related species on a single multiplex gene chip should be a practical, highly-parallel method for investigating genomic biodiversity.
Figures


References
-
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. - DOI - PubMed
-
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet. 2000;24:227–235. doi: 10.1038/73432. - DOI - PubMed
-
- Tiffin N, Adie E, Turner F, Brunner HG, van Driel MA, Oti M, Lopez-Bigas N, Ouzounis C, Perez-Iratxeta C, Andrade-Navarro MA, Adeyemo A, Patti ME, Semple CA, Hide W. Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nuc Acids Res. 2006;34:3067–3081. doi: 10.1093/nar/gkl381. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

