Systematic condition-dependent annotation of metabolic genes
- PMID: 17895423
- PMCID: PMC2045145
- DOI: 10.1101/gr.6678707
Systematic condition-dependent annotation of metabolic genes
Abstract
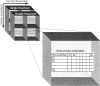
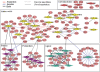
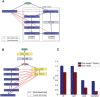
The task of deriving a functional annotation for genes is complex as their involvement in various processes depends on multiple factors such as environmental conditions and genetic backup mechanisms. This study employs a large-scale model of the metabolism of Saccharomyces cerevisiae to investigate the function of yeast genes and derive a condition-dependent annotation (CDA) for their involvement in major metabolic processes under various genetic and environmental conditions. The resulting CDA is validated on a large scale and is shown to be superior to the corresponding Gene Ontology (GO) annotation, by showing that genes annotated with the same CDA term tend to be more coherently conserved in evolution and display greater expression coherency than those annotated with the same GO term. The CDA gives rise to new kinds of functional condition-dependent metabolic pathways, some of which are described and further examined via substrate auxotrophy measurements of knocked-out strains. The CDA presented is likely to serve as a new reference source for metabolic gene annotation.
Figures

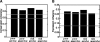
References
-
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. - PMC - PubMed
-
- Ball C.A., Awad I.A., Demeter J., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Awad I.A., Demeter J., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Demeter J., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Jin H., Matese J.C., Nitzberg M., Wymore F., Matese J.C., Nitzberg M., Wymore F., Nitzberg M., Wymore F., Wymore F., et al. The Stanford Microarray Database accommodates additional microarray platforms and data formats. Nucleic Acids Res. 2005;33:D580–D582. doi: 10.1093/nar/gki006. - DOI - PMC - PubMed
-
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C., Denouel A., Lacroute F., Cullin C., Lacroute F., Cullin C., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. - DOI - PMC - PubMed
-
- Brown J.A., Sherlock G., Myers C.L., Burrows N.M., Deng C., Wu H.I., McCann K.E., Troyanskaya O.G., Brown J.M., Sherlock G., Myers C.L., Burrows N.M., Deng C., Wu H.I., McCann K.E., Troyanskaya O.G., Brown J.M., Myers C.L., Burrows N.M., Deng C., Wu H.I., McCann K.E., Troyanskaya O.G., Brown J.M., Burrows N.M., Deng C., Wu H.I., McCann K.E., Troyanskaya O.G., Brown J.M., Deng C., Wu H.I., McCann K.E., Troyanskaya O.G., Brown J.M., Wu H.I., McCann K.E., Troyanskaya O.G., Brown J.M., McCann K.E., Troyanskaya O.G., Brown J.M., Troyanskaya O.G., Brown J.M., Brown J.M. Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100043. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
