Constraint-based modeling and kinetic analysis of the Smad dependent TGF-beta signaling pathway
- PMID: 17895977
- PMCID: PMC1978528
- DOI: 10.1371/journal.pone.0000936
Constraint-based modeling and kinetic analysis of the Smad dependent TGF-beta signaling pathway
Abstract
Background: Investigation of dynamics and regulation of the TGF-beta signaling pathway is central to the understanding of complex cellular processes such as growth, apoptosis, and differentiation. In this study, we aim at using systems biology approach to provide dynamic analysis on this pathway.
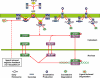
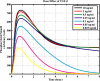
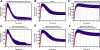
Methodology/principal findings: We proposed a constraint-based modeling method to build a comprehensive mathematical model for the Smad dependent TGF-beta signaling pathway by fitting the experimental data and incorporating the qualitative constraints from the experimental analysis. The performance of the model generated by constraint-based modeling method is significantly improved compared to the model obtained by only fitting the quantitative data. The model agrees well with the experimental analysis of TGF-beta pathway, such as the time course of nuclear phosphorylated Smad, the subcellular location of Smad and signal response of Smad phosphorylation to different doses of TGF-beta.
Conclusions/significance: The simulation results indicate that the signal response to TGF-beta is regulated by the balance between clathrin dependent endocytosis and non-clathrin mediated endocytosis. This model is useful to be built upon as new precise experimental data are emerging. The constraint-based modeling method can also be applied to quantitative modeling of other signaling pathways.
Conflict of interest statement
Figures



Similar articles
-
Distinct role of endocytosis for Smad and non-Smad TGF-β signaling regulation in hepatocytes.J Hepatol. 2011 Aug;55(2):369-78. doi: 10.1016/j.jhep.2010.11.027. Epub 2010 Dec 22. J Hepatol. 2011. PMID: 21184784
-
A mathematical model of the stoichiometric control of Smad complex formation in TGF-beta signal transduction pathway.J Theor Biol. 2009 Jul 21;259(2):389-403. doi: 10.1016/j.jtbi.2009.03.036. Epub 2009 Apr 7. J Theor Biol. 2009. PMID: 19358856
-
Cooperation of H2O2-mediated ERK activation with Smad pathway in TGF-beta1 induction of p21WAF1/Cip1.Cell Signal. 2006 Feb;18(2):236-43. doi: 10.1016/j.cellsig.2005.04.008. Epub 2005 Jun 24. Cell Signal. 2006. PMID: 15979845
-
TGF-beta signaling: a tale of two responses.J Cell Biochem. 2007 Oct 15;102(3):593-608. doi: 10.1002/jcb.21501. J Cell Biochem. 2007. PMID: 17729308 Review.
-
[TGF-beta/Smad in prostate cancer: an update].Zhonghua Nan Ke Xue. 2009 Sep;15(9):840-3. Zhonghua Nan Ke Xue. 2009. PMID: 19947572 Review. Chinese.
Cited by
-
Modeling the Control of TGF-β/Smad Nuclear Accumulation by the Hippo Pathway Effectors, Taz/Yap.iScience. 2020 Aug 21;23(8):101416. doi: 10.1016/j.isci.2020.101416. Epub 2020 Jul 29. iScience. 2020. PMID: 32798968 Free PMC article.
-
Colorectal cancer through simulation and experiment.IET Syst Biol. 2013 Jun;7(3):57-73. doi: 10.1049/iet-syb.2012.0019. IET Syst Biol. 2013. PMID: 24046975 Free PMC article. Review.
-
Multiscale cancer modeling.Annu Rev Biomed Eng. 2011 Aug 15;13:127-55. doi: 10.1146/annurev-bioeng-071910-124729. Annu Rev Biomed Eng. 2011. PMID: 21529163 Free PMC article. Review.
-
Dynamic regulation of Tgf-B signaling by Tif1γ: a computational approach.PLoS One. 2012;7(3):e33761. doi: 10.1371/journal.pone.0033761. Epub 2012 Mar 23. PLoS One. 2012. PMID: 22461896 Free PMC article.
-
Characterization of negative feedback network motifs in the TGF-β signaling pathway.PLoS One. 2013 Dec 20;8(12):e83531. doi: 10.1371/journal.pone.0083531. eCollection 2013. PLoS One. 2013. PMID: 24386222 Free PMC article.
References
-
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. - PubMed
-
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. - PubMed
-
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. - PubMed
-
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

