Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks
- PMID: 17895995
- PMCID: PMC1978522
- DOI: 10.1371/journal.pone.0000955
Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks
Abstract
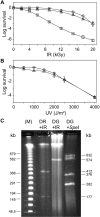
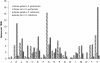
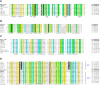
Bacteria of the genus Deinococcus are extremely resistant to ionizing radiation (IR), ultraviolet light (UV) and desiccation. The mesophile Deinococcus radiodurans was the first member of this group whose genome was completely sequenced. Analysis of the genome sequence of D. radiodurans, however, failed to identify unique DNA repair systems. To further delineate the genes underlying the resistance phenotypes, we report the whole-genome sequence of a second Deinococcus species, the thermophile Deinococcus geothermalis, which at its optimal growth temperature is as resistant to IR, UV and desiccation as D. radiodurans, and a comparative analysis of the two Deinococcus genomes. Many D. radiodurans genes previously implicated in resistance, but for which no sensitive phenotype was observed upon disruption, are absent in D. geothermalis. In contrast, most D. radiodurans genes whose mutants displayed a radiation-sensitive phenotype in D. radiodurans are conserved in D. geothermalis. Supporting the existence of a Deinococcus radiation response regulon, a common palindromic DNA motif was identified in a conserved set of genes associated with resistance, and a dedicated transcriptional regulator was predicted. We present the case that these two species evolved essentially the same diverse set of gene families, and that the extreme stress-resistance phenotypes of the Deinococcus lineage emerged progressively by amassing cell-cleaning systems from different sources, but not by acquisition of novel DNA repair systems. Our reconstruction of the genomic evolution of the Deinococcus-Thermus phylum indicates that the corresponding set of enzymes proliferated mainly in the common ancestor of Deinococcus. Results of the comparative analysis weaken the arguments for a role of higher-order chromosome alignment structures in resistance; more clearly define and substantially revise downward the number of uncharacterized genes that might participate in DNA repair and contribute to resistance; and strengthen the case for a role in survival of systems involved in manganese and iron homeostasis.
Conflict of interest statement
Figures


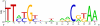
References
-
- Lai WA, Kampfer P, Arun AB, Shen FT, Huber B, et al. Deinococcus ficus sp. nov., isolated from the rhizosphere of Ficus religiosa L. Int J Syst Evol Microbiol. 2006;56:787–791. - PubMed
-
- Gutman PD, Fuchs P, Minton KW. Restoration of the DNA damage resistance of Deinococcus radiodurans DNA polymerase mutants by Escherichia coli DNA polymerase I and Klenow fragment. Mutat Res. 1994;314:87–97. - PubMed
-
- Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

