pHUSH: a single vector system for conditional gene expression
- PMID: 17897455
- PMCID: PMC2174931
- DOI: 10.1186/1472-6750-7-61
pHUSH: a single vector system for conditional gene expression
Abstract
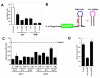
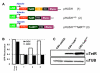
Background: Conditional expression vectors have become a valuable research tool to avoid artefacts that may result from traditional gene expression studies. However, most systems require multiple plasmids that must be independently engineered into the target system, resulting in experimental delay and an increased potential for selection of a cell subpopulation that differs significantly from the parental line. We have therefore developed pHUSH, an inducible expression system that allows regulated expression of shRNA, miRNA or cDNA cassettes on a single viral vector.
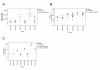
Results: Both Pol II and Pol III promoters have been successfully combined with a second expression cassette containing a codon-optimized tetracycline repressor and selectable marker. We provide examples of how pHUSH has been successfully employed to study the function of target genes in a number of cell types within in vitro and in vivo assays, including conditional gene knockdown in a murine model of brain cancer.
Conclusion: We have successfully developed and employed a single vector system that enables Doxycycline regulated RNAi or transgene expression in a variety of in vitro and in vivo model systems. These studies demonstrate the broad application potential of pHUSH for conditional genetic engineering in mammalian cells.
Figures
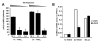


References
-
- Sandy P, Ventura A, Jacks T. Mammalian RNAi: a practical guide. Biotechniques. 2005;39:215–224. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

