Ras nanoclusters: molecular structure and assembly
- PMID: 17897845
- PMCID: PMC2761225
- DOI: 10.1016/j.semcdb.2007.08.003
Ras nanoclusters: molecular structure and assembly
Abstract
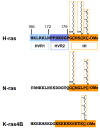
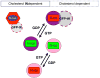
H-, N- and K-ras4B are lipid-anchored, peripheral membrane guanine nucleotide binding proteins. Recent work has shown that Ras proteins are laterally segregated into non-overlapping, dynamic domains of the plasma membrane called nanoclusters. This lateral segregation is important to specify Ras interactions with membrane-associated proteins, effectors and scaffolding proteins and is critical for Ras signal transduction. Here we review biological, in vitro and structural data that provide insight into the molecular basis of how palmitoylated Ras proteins are anchored to the plasma membrane. We explore possible mechanisms for how the interactions of H-ras with a lipid bilayer may drive nanocluster formation.
Figures



References
-
- Hancock JF, Magee AI, Childs JE, Marshall CJ. Cell. 1989;57:1167–77. - PubMed
-
- Bergo MO, Lieu HD, Gavino BJ, Ambroziak P, Otto JC, Casey PJ, Walker QM, Young SG. J Biol Chem. 2004;279:4729–36. - PubMed
-
- Pillinger MH, Volker C, Stock JB, Weissmann G, Philips MR. J Biol Chem. 1994;269:1486–92. - PubMed
-
- Hancock JF. Nat Rev Mol Cell Biol. 2003;4:373–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

