Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation
- PMID: 17898715
- PMCID: PMC3350864
- DOI: 10.1038/nature06160
Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation
Abstract
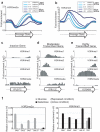
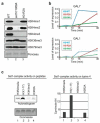
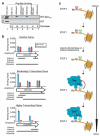
Modifications on histones control important biological processes through their effects on chromatin structure. Methylation at lysine 4 on histone H3 (H3K4) is found at the 5' end of active genes and contributes to transcriptional activation by recruiting chromatin-remodelling enzymes. An adjacent arginine residue (H3R2) is also known to be asymmetrically dimethylated (H3R2me2a) in mammalian cells, but its location within genes and its function in transcription are unknown. Here we show that H3R2 is also methylated in budding yeast (Saccharomyces cerevisiae), and by using an antibody specific for H3R2me2a in a chromatin immunoprecipitation-on-chip analysis we determine the distribution of this modification on the entire yeast genome. We find that H3R2me2a is enriched throughout all heterochromatic loci and inactive euchromatic genes and is present at the 3' end of moderately transcribed genes. In all cases the pattern of H3R2 methylation is mutually exclusive with the trimethyl form of H3K4 (H3K4me3). We show that methylation at H3R2 abrogates the trimethylation of H3K4 by the Set1 methyltransferase. The specific effect on H3K4me3 results from the occlusion of Spp1, a Set1 methyltransferase subunit necessary for trimethylation. Thus, the inability of Spp1 to recognize H3 methylated at R2 prevents Set1 from trimethylating H3K4. These results provide the first mechanistic insight into the function of arginine methylation on chromatin.
Figures

References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. - PubMed
-
- Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–55. - PubMed
-
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. - PubMed
-
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–86. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

