High variability and non-neutral evolution of the mammalian avpr1a gene
- PMID: 17900345
- PMCID: PMC2121647
- DOI: 10.1186/1471-2148-7-176
High variability and non-neutral evolution of the mammalian avpr1a gene
Abstract
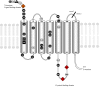
Background: The arginine-vasopressin 1a receptor has been identified as a key determinant for social behaviour in Microtus voles, humans and other mammals. Nevertheless, the genetic bases of complex phenotypic traits like differences in social and mating behaviour among species and individuals remain largely unknown. Contrary to previous studies focusing on differences in the promotor region of the gene, we investigate here the level of functional variation in the coding region (exon 1) of this locus.
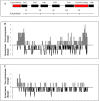
Results: We detected high sequence diversity between higher mammalian taxa as well as between species of the genus Microtus. This includes length variation and radical amino acid changes, as well as the presence of distinct protein variants within individuals. Additionally, negative selection prevails on most parts of the first exon of the arginine-vasopressin receptor 1a (avpr1a) gene but it contains regions with higher rates of change that harbour positively selected sites. Synonymous and non-synonymous substitution rates in the avpr1a gene are not exceptional compared to other genes, but they exceed those found in related hormone receptors with similar functions.
Discussion: These results stress the importance of considering variation in the coding sequence of avpr1a in regards to associations with life history traits (e.g. social behaviour, mating system, habitat requirements) of voles, other mammals and humans in particular.
Figures



Similar articles
-
Evolution of the arginine vasopressin 1a receptor and implications for mammalian social behaviour.Prog Brain Res. 2008;170:321-30. doi: 10.1016/S0079-6123(08)00426-3. Prog Brain Res. 2008. PMID: 18655892
-
Complex selection on a regulator of social cognition: Evidence of balancing selection, regulatory interactions and population differentiation in the prairie vole Avpr1a locus.Mol Ecol. 2018 Jan;27(2):419-431. doi: 10.1111/mec.14455. Epub 2017 Dec 29. Mol Ecol. 2018. PMID: 29218792
-
Polymorphism at the avpr1a locus in male prairie voles correlated with genetic but not social monogamy in field populations.Mol Ecol. 2009 Nov;18(22):4680-95. doi: 10.1111/j.1365-294X.2009.04361.x. Epub 2009 Oct 12. Mol Ecol. 2009. PMID: 19821904
-
Molecular genetic studies of the arginine vasopressin 1a receptor (AVPR1a) and the oxytocin receptor (OXTR) in human behaviour: from autism to altruism with some notes in between.Prog Brain Res. 2008;170:435-49. doi: 10.1016/S0079-6123(08)00434-2. Prog Brain Res. 2008. PMID: 18655900 Review.
-
Gene regulation as a modulator of social preference in voles.Adv Genet. 2007;59:107-27. doi: 10.1016/S0065-2660(07)59004-8. Adv Genet. 2007. PMID: 17888796 Review.
Cited by
-
Sex-specific clines support incipient speciation in a common European mammal.Heredity (Edinb). 2013 Apr;110(4):398-404. doi: 10.1038/hdy.2012.124. Epub 2013 Jan 23. Heredity (Edinb). 2013. PMID: 23340600 Free PMC article.
-
Increased expression and protein divergence in duplicate genes is associated with morphological diversification.PLoS Genet. 2009 Dec;5(12):e1000781. doi: 10.1371/journal.pgen.1000781. Epub 2009 Dec 24. PLoS Genet. 2009. PMID: 20041196 Free PMC article.
-
Music genetics research: Association with musicality of a polymorphism in the AVPR1A gene.Genet Mol Biol. 2017 Apr-Jun;40(2):421-429. doi: 10.1590/1678-4685-GMB-2016-0021. Epub 2017 May 22. Genet Mol Biol. 2017. PMID: 28534928 Free PMC article.
-
A complex selection signature at the human AVPR1B gene.BMC Evol Biol. 2009 Jun 1;9:123. doi: 10.1186/1471-2148-9-123. BMC Evol Biol. 2009. PMID: 19486526 Free PMC article.
-
Gender-specific Single Transcript Level Atlas of Vasopressin and its Receptor (AVPR1a) in the Mouse Brain.bioRxiv [Preprint]. 2024 Dec 10:2024.12.09.627541. doi: 10.1101/2024.12.09.627541. bioRxiv. 2024. PMID: 39713333 Free PMC article. Preprint.
References
-
- Lank DB, Smith CM, Hanotte O, Burke T, Cooke F. Genetic polymorphism for alternative mating behaviour in lekking male ruff Philomachus pugnax. Nature. 1995;378:59–62. doi: 10.1038/378059a0. - DOI
-
- Shuster SM, Sassaman C. Genetic interaction between male mating strategy and sex ratio in a marine isopod. Nature. 1997;388:373–377. doi: 10.1038/41089. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

