Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes
- PMID: 17900370
- PMCID: PMC2121648
- DOI: 10.1186/1471-2148-7-178
Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes
Abstract
Background: The reactive oxygen-generating NADPH oxidases (Noxes) function in a variety of biological roles, and can be broadly classified into those that are regulated by subunit interactions and those that are regulated by calcium. The prototypical subunit-regulated Nox, Nox2, is the membrane-associated catalytic subunit of the phagocyte NADPH-oxidase. Nox2 forms a heterodimer with the integral membrane protein, p22phox, and this heterodimer binds to the regulatory subunits p47phox, p67phox, p40phox and the small GTPase Rac, triggering superoxide generation. Nox-organizer protein 1 (NOXO1) and Nox-activator 1 (NOXA1), respective homologs of p47phox and p67phox, together with p22phox and Rac, activate Nox1, a non-phagocytic homolog of Nox2. NOXO1 and p22phox also regulate Nox3, whereas Nox4 requires only p22phox. In this study, we have assembled and analyzed amino acid sequences of Nox regulatory subunit orthologs from vertebrates, a urochordate, an echinoderm, a mollusc, a cnidarian, a choanoflagellate, fungi and a slime mold amoeba to investigate the evolutionary history of these subunits.
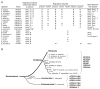
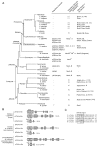
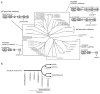
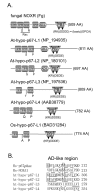
Results: Ancestral p47phox, p67phox, and p22phox genes are broadly seen in the metazoa, except for the ecdysozoans. The choanoflagellate Monosiga brevicollis, the unicellular organism that is the closest relatives of multicellular animals, encodes early prototypes of p22phox, p47phox as well as the earliest known Nox2-like ancestor of the Nox1-3 subfamily. p67phox- and p47phox-like genes are seen in the sea urchin Strongylocentrotus purpuratus and the limpet Lottia gigantea that also possess Nox2-like co-orthologs of vertebrate Nox1-3. Duplication of primordial p47phox and p67phox genes occurred in vertebrates, with the duplicated branches evolving into NOXO1 and NOXA1. Analysis of characteristic domains of regulatory subunits suggests a novel view of the evolution of Nox: in fish, p40phox participated in regulating both Nox1 and Nox2, but after the appearance of mammals, Nox1 (but not Nox2) became independent of p40phox. In the fish Oryzias latipes, a NOXO1 ortholog retains an autoinhibitory region that is characteristic of mammalian p47phox, and this was subsequently lost from NOXO1 in later vertebrates. Detailed amino acid sequence comparisons identified both putative key residues conserved in characteristic domains and previously unidentified conserved regions. Also, candidate organizer/activator proteins in fungi and amoeba are identified and hypothetical activation models are suggested.
Conclusion: This is the first report to provide the comprehensive view of the molecular evolution of regulatory subunits for Nox enzymes. This approach provides clues for understanding the evolution of biochemical and physiological functions for regulatory-subunit-dependent Nox enzymes.
Figures


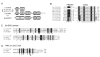

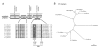
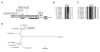




Similar articles
-
Point mutations in the proline-rich region of p22phox are dominant inhibitors of Nox1- and Nox2-dependent reactive oxygen generation.J Biol Chem. 2005 Sep 9;280(36):31859-69. doi: 10.1074/jbc.M501882200. Epub 2005 Jun 30. J Biol Chem. 2005. PMID: 15994299
-
Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes.BMC Evol Biol. 2007 Jul 6;7:109. doi: 10.1186/1471-2148-7-109. BMC Evol Biol. 2007. PMID: 17612411 Free PMC article.
-
Soluble Regulatory Proteins for Activation of NOX Family NADPH Oxidases.Methods Mol Biol. 2019;1982:121-137. doi: 10.1007/978-1-4939-9424-3_8. Methods Mol Biol. 2019. PMID: 31172470
-
Role of the small GTPase Rac in p22phox-dependent NADPH oxidases.Biochimie. 2007 Sep;89(9):1133-44. doi: 10.1016/j.biochi.2007.05.003. Epub 2007 May 17. Biochimie. 2007. PMID: 17583407 Review.
-
Organizers and activators: Cytosolic Nox proteins impacting on vascular function.Free Radic Biol Med. 2017 Aug;109:22-32. doi: 10.1016/j.freeradbiomed.2017.03.017. Epub 2017 Mar 21. Free Radic Biol Med. 2017. PMID: 28336130 Review.
Cited by
-
Cell signaling through protein kinase C oxidation and activation.Int J Mol Sci. 2012;13(9):10697-10721. doi: 10.3390/ijms130910697. Epub 2012 Aug 24. Int J Mol Sci. 2012. PMID: 23109817 Free PMC article. Review.
-
A conserved region between the TPR and activation domains of p67phox participates in activation of the phagocyte NADPH oxidase.J Biol Chem. 2010 Oct 8;285(41):31435-45. doi: 10.1074/jbc.M110.161166. Epub 2010 Aug 2. J Biol Chem. 2010. PMID: 20679349 Free PMC article.
-
In silico and gene expression analysis of the acute inflammatory response of gilthead seabream (Sparus aurata) after subcutaneous administration of carrageenin.Fish Physiol Biochem. 2021 Oct;47(5):1623-1643. doi: 10.1007/s10695-021-00999-6. Epub 2021 Aug 26. Fish Physiol Biochem. 2021. PMID: 34448108 Free PMC article.
-
Nox5 forms a functional oligomer mediated by self-association of its dehydrogenase domain.Biochemistry. 2011 Mar 29;50(12):2013-25. doi: 10.1021/bi1020088. Epub 2011 Mar 4. Biochemistry. 2011. PMID: 21319793 Free PMC article.
-
Phosphatidylinositol (4,5)-bisphosphate modulates Nox5 localization via an N-terminal polybasic region.Mol Biol Cell. 2008 Oct;19(10):4020-31. doi: 10.1091/mbc.e07-12-1223. Epub 2008 Jul 9. Mol Biol Cell. 2008. PMID: 18614798 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

