Paired-end mapping reveals extensive structural variation in the human genome
- PMID: 17901297
- PMCID: PMC2674581
- DOI: 10.1126/science.1149504
Paired-end mapping reveals extensive structural variation in the human genome
Abstract
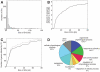
Structural variation of the genome involves kilobase- to megabase-sized deletions, duplications, insertions, inversions, and complex combinations of rearrangements. We introduce high-throughput and massive paired-end mapping (PEM), a large-scale genome-sequencing method to identify structural variants (SVs) approximately 3 kilobases (kb) or larger that combines the rescue and capture of paired ends of 3-kb fragments, massive 454 sequencing, and a computational approach to map DNA reads onto a reference genome. PEM was used to map SVs in an African and in a putatively European individual and identified shared and divergent SVs relative to the reference genome. Overall, we fine-mapped more than 1000 SVs and documented that the number of SVs among humans is much larger than initially hypothesized; many of the SVs potentially affect gene function. The breakpoint junction sequences of more than 200 SVs were determined with a novel pooling strategy and computational analysis. Our analysis provided insights into the mechanisms of SV formation in humans.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

