The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA
- PMID: 17905813
- PMCID: PMC2095795
- DOI: 10.1093/nar/gkm741
The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA
Abstract
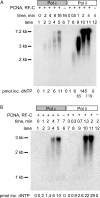
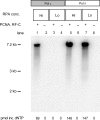
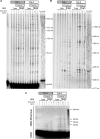
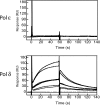
Saccharomyces cerevisiae DNA polymerase delta (Pol delta) and DNA polymerase epsilon (Pol epsilon) are replicative DNA polymerases at the replication fork. Both enzymes are stimulated by PCNA, although to different levels. To understand why and to explore the interaction with PCNA, we compared Pol delta and Pol epsilon in physical interactions with PCNA and nucleic acids (with or without RPA), and in functional assays measuring activity and processivity. Using surface plasmon resonance technique, we show that Pol epsilon has a high affinity for DNA, but a low affinity for PCNA. In contrast, Pol delta has a low affinity for DNA and a high affinity for PCNA. The true processivity of Pol delta and Pol epsilon was measured for the first time in the presence of RPA, PCNA and RFC on single-stranded DNA. Remarkably, in the presence of PCNA, the processivity of Pol delta and Pol epsilon on RPA-coated DNA is comparable. Finally, more PCNA molecules were found on the template after it was replicated by Pol epsilon when compared to Pol delta. We conclude that Pol epsilon and Pol delta exhibit comparable processivity, but are loaded on the primer-end via different mechanisms.
Figures



References
-
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. - PubMed
-
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. - PubMed
-
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

