Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots
- PMID: 17905859
- PMCID: PMC2048793
- DOI: 10.1104/pp.107.104844
Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots
Abstract
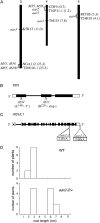
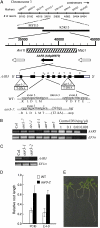
To isolate novel auxin-responsive mutants in Arabidopsis (Arabidopsis thaliana), we screened mutants for root growth resistance to a putative antiauxin, p-chlorophenoxyisobutyric acid (PCIB), which inhibits auxin action by interfering the upstream auxin-signaling events. Eleven PCIB-resistant mutants were obtained. Genetic mapping indicates that the mutations are located in at least five independent loci, including two known auxin-related loci, TRANSPORT INHIBITOR RESPONSE1 and Arabidopsis CULLIN1. antiauxin-resistant mutants (aars) aar3-1, aar4, and aar5 were also resistant to 2,4-dichlorophenoxyacetic acid as shown by a root growth assay. Positional cloning of aar3-1 revealed that the AAR3 gene encodes a protein with a domain of unknown function (DUF298), which has not previously been implicated in auxin signaling. The protein has a putative nuclear localization signal and shares homology with the DEFECTIVE IN CULLIN NEDDYLATION-1 protein through the DUF298 domain. The results also indicate that PCIB can facilitate the identification of factors involved in auxin or auxin-related signaling.
Figures




References
-
- Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8 87–96 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Biswas KK, Neumann R, Haga K, Yatoh O, Iino M (2003) Photomorphogenesis of rice seedlings: a mutant impaired in phytochrome-mediated inhibition of coleoptile growth. Plant Cell Physiol 44 242–254 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

