Global expression profiling applied to the analysis of Arabidopsis stamen development
- PMID: 17905860
- PMCID: PMC2048804
- DOI: 10.1104/pp.107.104422
Global expression profiling applied to the analysis of Arabidopsis stamen development
Abstract
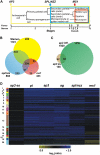
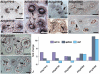
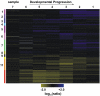
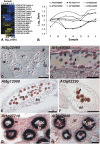
To obtain detailed information about gene expression during stamen development in Arabidopsis (Arabidopsis thaliana), we compared, by microarray analysis, the gene expression profile of wild-type inflorescences to those of the floral mutants apetala3, sporocyteless/nozzle, and male sterile1 (ms1), in which different aspects of stamen formation are disrupted. These experiments led to the identification of groups of genes with predicted expression at early, intermediate, and late stages of stamen development. Validation experiments using in situ hybridization confirmed the predicted expression patterns. Additional experiments aimed at characterizing gene expression specifically during microspore formation. To this end, we compared the gene expression profiles of wild-type flowers of distinct developmental stages to those of the ms1 mutant. Computational analysis of the datasets derived from this experiment led to the identification of genes that are likely involved in the control of key developmental processes during microsporogenesis. We also identified a large number of genes whose expression is prolonged in ms1 mutant flowers compared to the wild type. This result suggests that MS1, which encodes a putative transcriptional regulator, is involved in the stage-specific repression of these genes. Lastly, we applied reverse genetics to characterize several of the genes identified in the microarray experiments and uncovered novel regulators of microsporogenesis, including the transcription factor MYB99 and a putative phosphatidylinositol 4-kinase.
Figures


References
-
- Aarts MGM, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12 615–623 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Amagai M, Ariizumi T, Endo M, Hatakeyama K, Kuwata C, Shibata D, Toriyama K, Watanabe M (2003) Identification of anther-specific genes in a cruciferous model plant, Arabidopsis thaliana, by using a combination of Arabidopsis macroarray and mRNA derived from Brassica oleracea. Sex Plant Reprod 15 213–220
-
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K (2005) The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sex Plant Reprod 18 1–7
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

