The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses
- PMID: 17905899
- PMCID: PMC2048692
- DOI: 10.1105/tpc.106.049981
The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses
Abstract
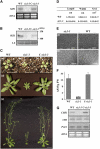
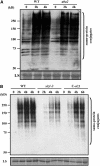
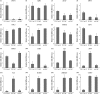
Posttranslational modifications of proteins by small ubiquitin-like modifiers (SUMOs) regulate protein degradation and localization, protein-protein interaction, and transcriptional activity. SUMO E3 ligase functions are executed by SIZ1/SIZ2 and Mms21 in yeast, the PIAS family members RanBP2, and Pc2 in human. The Arabidopsis thaliana genome contains only one gene, SIZ1, that is orthologous to the yeast SIZ1/SIZ2. Here, we show that Arabidopsis SIZ1 is expressed in all plant tissues. Compared with the wild type, the null mutant siz1-3 is smaller in stature because of reduced expression of genes involved in brassinosteroid biosynthesis and signaling. Drought stress induces the accumulation of SUMO-protein conjugates, which is in part dependent on SIZ1 but not on abscisic acid (ABA). Mutant plants of siz1-3 have significantly lower tolerance to drought stress. A genome-wide expression analysis identified approximately 1700 Arabidopsis genes that are induced by drought, with SIZ1 mediating the expression of 300 of them by a pathway independent of DREB2A and ABA. SIZ1-dependent, drought-responsive genes include those encoding enzymes of the anthocyanin synthesis pathway and jasmonate response. From these results, we conclude that SIZ1 regulates Arabidopsis growth and that this SUMO E3 ligase plays a role in drought stress response likely through the regulation of gene expression.
Figures


Similar articles
-
Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance.Plant Physiol. 2011 Aug;156(4):2225-34. doi: 10.1104/pp.111.178996. Epub 2011 Jun 1. Plant Physiol. 2011. PMID: 21632972 Free PMC article.
-
SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F).J Integr Plant Biol. 2013 Jan;55(1):83-95. doi: 10.1111/jipb.12024. J Integr Plant Biol. 2013. PMID: 23231763
-
SIZ1-Dependent Post-Translational Modification by SUMO Modulates Sugar Signaling and Metabolism in Arabidopsis thaliana.Plant Cell Physiol. 2015 Dec;56(12):2297-311. doi: 10.1093/pcp/pcv149. Epub 2015 Oct 14. Plant Cell Physiol. 2015. PMID: 26468507
-
A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco.J Integr Plant Biol. 2017 Feb;59(2):102-117. doi: 10.1111/jipb.12514. J Integr Plant Biol. 2017. PMID: 27995772
-
Cytoplasmic sumoylation by PIAS-type Siz1-SUMO ligase.Cell Cycle. 2008 Jun 15;7(12):1738-44. doi: 10.4161/cc.7.12.6156. Epub 2008 Jun 16. Cell Cycle. 2008. PMID: 18583943 Review.
Cited by
-
A non-invasive method to predict drought survival in Arabidopsis using quantum yield under light conditions.Plant Methods. 2023 Nov 15;19(1):127. doi: 10.1186/s13007-023-01107-w. Plant Methods. 2023. PMID: 37968652 Free PMC article.
-
Insights into the transcriptional and post-transcriptional regulation of the rice SUMOylation machinery and into the role of two rice SUMO proteases.BMC Plant Biol. 2018 Dec 12;18(1):349. doi: 10.1186/s12870-018-1547-3. BMC Plant Biol. 2018. PMID: 30541427 Free PMC article.
-
PHD-finger family genes in wheat (Triticum aestivum L.): Evolutionary conservatism, functional diversification, and active expression in abiotic stress.Front Plant Sci. 2022 Dec 12;13:1016831. doi: 10.3389/fpls.2022.1016831. eCollection 2022. Front Plant Sci. 2022. PMID: 36578331 Free PMC article.
-
Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection.J Virol. 2011 Oct;85(19):9789-800. doi: 10.1128/JVI.02566-10. Epub 2011 Jul 20. J Virol. 2011. PMID: 21775461 Free PMC article.
-
Integration of low temperature and light signaling during cold acclimation response in Arabidopsis.Proc Natl Acad Sci U S A. 2011 Sep 27;108(39):16475-80. doi: 10.1073/pnas.1107161108. Epub 2011 Sep 19. Proc Natl Acad Sci U S A. 2011. PMID: 21930922 Free PMC article.
References
-
- Arteca, R.N., Bachman, J.M., Tsai, D.S., and Mandava, N.B. (1988). Fusiccocin, an inhibitor of brassinosteroid-induced ethylene production. Physiol. Plant. 74 631–634.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

