Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation
- PMID: 17905924
- PMCID: PMC2168408
- DOI: 10.1128/EC.00285-07
Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation
Abstract
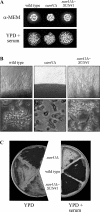
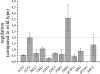
The SUN gene family has been defined in Saccharomyces cerevisiae and comprises a fungus-specific family of proteins which show high similarity in their C-terminal domains. Genes of this family are involved in different cellular processes, like DNA replication, aging, mitochondrial biogenesis, and cytokinesis. In Candida albicans the SUN family comprises two genes, SUN41 and SIM1. We demonstrate that C. albicans mutants lacking SUN41 show similar defects as found for S. cerevisiae, including defects in cytokinesis. In addition, the SUN41 mutant showed a higher sensitivity towards the cell wall-disturbing agent Congo red, whereas no difference was observed in the presence of calcofluor white. Compared to the wild type, SUN41 deletion strains exhibited a defect in biofilm formation, a reduced adherence on a Caco-2 cell monolayer, and were unable to form hyphae on solid medium under the conditions tested. Interestingly, Sun41p was found to be secreted in the medium of cells growing as blastospores as well as those forming hyphae. Our results support a function of SUN41p as a glycosidase involved in cytokinesis, cell wall biogenesis, adhesion to host tissue, and biofilm formation, indicating an important role in the host-pathogen interaction.
Figures





Similar articles
-
SUN proteins belong to a novel family of β-(1,3)-glucan-modifying enzymes involved in fungal morphogenesis.J Biol Chem. 2013 May 10;288(19):13387-96. doi: 10.1074/jbc.M112.440172. Epub 2013 Mar 18. J Biol Chem. 2013. PMID: 23508952 Free PMC article.
-
The SUN41 and SUN42 genes are essential for cell separation in Candida albicans.Mol Microbiol. 2007 Dec;66(5):1256-75. doi: 10.1111/j.1365-2958.2007.06011.x. Mol Microbiol. 2007. PMID: 18001349
-
Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms.Eukaryot Cell. 2013 Feb;12(2):224-32. doi: 10.1128/EC.00163-12. Epub 2012 Dec 7. Eukaryot Cell. 2013. PMID: 23223035 Free PMC article.
-
Involvement of amyloid proteins in the formation of biofilms in the pathogenic yeast Candida albicans.Res Microbiol. 2021 Apr-May;172(3):103813. doi: 10.1016/j.resmic.2021.103813. Epub 2021 Jan 28. Res Microbiol. 2021. PMID: 33515679 Review.
-
Candida albicans cell wall proteins.Microbiol Mol Biol Rev. 2008 Sep;72(3):495-544. doi: 10.1128/MMBR.00032-07. Microbiol Mol Biol Rev. 2008. PMID: 18772287 Free PMC article. Review.
Cited by
-
Complementary adhesin function in C. albicans biofilm formation.Curr Biol. 2008 Jul 22;18(14):1017-24. doi: 10.1016/j.cub.2008.06.034. Curr Biol. 2008. PMID: 18635358 Free PMC article.
-
The APSES transcription factor Vst1 is a key regulator of development in microsclerotium- and resting mycelium-producing Verticillium species.Mol Plant Pathol. 2018 Jan;19(1):59-76. doi: 10.1111/mpp.12496. Epub 2017 Jan 13. Mol Plant Pathol. 2018. PMID: 27696683 Free PMC article.
-
Argonaute and Dicer are essential for communication between Trichoderma atroviride and fungal hosts during mycoparasitism.Microbiol Spectr. 2024 Apr 2;12(4):e0316523. doi: 10.1128/spectrum.03165-23. Epub 2024 Mar 5. Microbiol Spectr. 2024. PMID: 38441469 Free PMC article.
-
Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans.Fungal Genet Biol. 2009 Oct;46(10):768-81. doi: 10.1016/j.fgb.2009.06.005. Epub 2009 Jun 23. Fungal Genet Biol. 2009. PMID: 19555771 Free PMC article.
-
The Saccharomyces SUN gene, UTH1, is involved in cell wall biogenesis.FEMS Yeast Res. 2010 Mar;10(2):168-76. doi: 10.1111/j.1567-1364.2009.00601.x. Epub 2009 Dec 18. FEMS Yeast Res. 2010. PMID: 20070376 Free PMC article.
References
-
- Al-Fattani, M. A., and L. J. Douglas. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J. Med. Microbiol. 55:999-1008. - PubMed
-
- Camougrand, N. M., M. Mouassite, G. M. Velours, and M. G. Guerin. 2000. The “SUN” family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 375:154-160. - PubMed
-
- Dieterich, C., M. Schandar, M. Noll, F. J. Johannes, H. Brunner, T. Graeve, and S. Rupp. 2002. In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology 148:497-506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

