The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the Group A streptococcus
- PMID: 17905980
- PMCID: PMC2168945
- DOI: 10.1128/JB.01038-07
The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the Group A streptococcus
Abstract
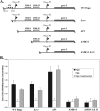
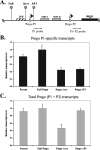
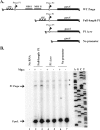
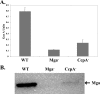
Carbon catabolite repression (CCR) allows bacteria to alter metabolism in response to the availability of specific sugar sources, and increasing evidence suggests that CCR is involved in regulating virulence gene expression in many pathogens. A scan of the M1 SF370 group A streptococcus (GAS) genome using a Bacillus subtilis consensus identified a number of potential catabolite-responsive elements (cre) important for binding by the catabolite control protein A (CcpA), a mediator of CCR in gram-positive bacteria. Intriguingly, a putative cre was identified in the promoter region of mga upstream of its distal P1 start of transcription. Electrophoretic mobility shift assays showed that a His-CcpA fusion protein was capable of binding specifically to the cre in Pmga in vitro. Deletion analysis of Pmga using single-copy Pmga-gusA reporter strains found that Pmga P1 and its upstream cre were not required for normal autoregulated mga expression from Pmga P2 as long as Mga was produced from its native locus. In fact, the Pmga P1 region appeared to show a negative influence on Pmga P2 in these studies. However, deletion of the cre at the native Pmga resulted in a reduction of total mga transcripts as determined by real-time reverse transcription-PCR, supporting a role for CcpA in initial expression. Furthermore, normal transcriptional initiation from the Pmga P1 start site alone was dependent on the presence of the cre. Importantly, inactivation of ccpA in the M6 GAS strain JRS4 resulted in a reduction in Pmga expression and Mga protein levels in late-logarithmic-phase cell growth. These data support a role for CcpA in the early activation of the mga promoter and establish a link between CCR and Mga regulation in the GAS.
Figures


References
-
- Aung-Hilbrich, L. M., G. Seidel, A. Wagner, and W. Hillen. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J. Mol. Biol. 319:77-85. - PubMed
-
- Deutscher, J., E. Kuster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15:1049-1053. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

