Distribution of centromere-like parS sites in bacteria: insights from comparative genomics
- PMID: 17905987
- PMCID: PMC2168934
- DOI: 10.1128/JB.01239-07
Distribution of centromere-like parS sites in bacteria: insights from comparative genomics
Abstract
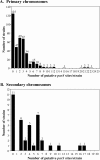
Partitioning of low-copy-number plasmids to daughter cells often depends on ParA and ParB proteins acting on centromere-like parS sites. Similar chromosome-encoded par loci likely also contribute to chromosome segregation. Here, we used bioinformatic approaches to search for chromosomal parS sites in 400 prokaryotic genomes. Although the consensus sequence matrix used to search for parS sites was derived from two gram-positive species, putative parS sites were identified on the chromosomes of 69% of strains from all branches of bacteria. Strains that were not found to contain parS sites clustered among relatively few branches of the prokaryotic evolutionary tree. In the vast majority of cases, parS sites were identified in origin-proximal regions of chromosomes. The widespread conservation of parS sites across diverse bacteria suggests that par loci evolved very early in the evolution of bacterial chromosomes and that the absence of parS, parA, and/or parB in certain strains likely reflects the loss of one of more of these loci much later in evolution. Moreover, the highly conserved origin-proximal position of parS suggests par loci are primarily devoted to regulating processes that involve the origin region of bacterial chromosomes. In species containing multiple chromosomes, the parS sites found on secondary chromosomes diverge significantly from those found on their primary chromosomes, suggesting that chromosome segregation of multipartite genomes requires distinct replicon-specific par loci. Furthermore, parS sites on secondary chromosomes are not well conserved among different species, suggesting that the evolutionary histories of secondary chromosomes are more diverse than those of primary chromosomes.
Figures




References
-
- Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

