Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains
- PMID: 17905991
- PMCID: PMC2168937
- DOI: 10.1128/JB.01150-07
Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains
Erratum in
- J Bacteriol. 2008 May;190(10):3791
Abstract
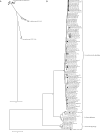
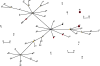
The population genetic structure of the animal pathogen Staphylococcus intermedius is poorly understood. We carried out a multilocus sequence phylogenetic analysis of isolates from broad host and geographic origins to investigate inter- and intraspecies diversity. We found that isolates phenotypically identified as S. intermedius are differentiated into three closely related species, S. intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini. S. pseudintermedius, not S. intermedius, is the common cause of canine pyoderma and occasionally causes zoonotic infections of humans. Over 60 extant STs were identified among the S. pseudintermedius isolates examined, including several that were distributed on different continents. As the agr quorum-sensing system of staphylococci is thought to have evolved along lines of speciation within the genus, we examined the allelic variation of agrD, which encodes the autoinducing peptide (AIP). Four AIP variants were encoded by S. pseudintermedius isolates, and identical AIP variants were shared among the three species, suggesting that a common quorum-sensing capacity has been conserved in spite of species differentiation in largely distinct ecological niches. A lack of clonal association of agr alleles suggests that assortive recombination may have contributed to the distribution of agr diversity. Finally, we discovered that the recent emergence of methicillin-resistant strains was due to multiple acquisitions of the mecA gene by different S. pseudintermedius clones found on different continents. Taken together, these data have resolved the population genetic structure of the S. intermedius group, resulting in new insights into its ancient and recent evolution.
Figures
References
-
- Aarestrup, F. M. 2001. Comparative ribotyping of Staphylococcus intermedius isolated from members of the Canoidea gives possible evidence for host-specificity and co-evolution of bacteria and hosts. Int. J. Syst. Evol. Microbiol. 51:1343-1347. - PubMed
-
- Bes, M., L. Saidi Slim, F. Becharnia, H. Meugnier, F. Vandenesch, J. Etienne, and J. Freney. 2002. Population diversity of Staphylococcus intermedius isolates from various host species: typing by 16S-23S intergenic ribosomal DNA spacer polymorphism analysis. J. Clin. Microbiol. 40:2275-2277. - PMC - PubMed
-
- Chesneau, O., A. Morvan, S. Aubert, and N. el Solh. 2000. The value of rRNA gene restriction site polymorphism analysis for delineating taxa in the genus Staphylococcus. Int. J. Syst. Evol. Microbiol. 50:689-697. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

