Insight into the haem d1 biosynthesis pathway in heliobacteria through bioinformatics analysis
- PMID: 17906152
- PMCID: PMC2774728
- DOI: 10.1099/mic.0.2007/007930-0
Insight into the haem d1 biosynthesis pathway in heliobacteria through bioinformatics analysis
Abstract
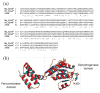
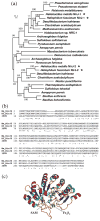
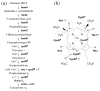
Haem d(1) is a unique tetrapyrrole molecule that serves as a prosthetic group of cytochrome cd(1), which reduces nitrite to nitric oxide during the process of denitrification. Very little information is available regarding the biosynthesis of haem d(1). The extreme difficulty in studying the haem d(1) biosynthetic pathway can be partly attributed to the lack of a theoretical basis for experimental investigation. We report here a gene cluster encoding enzymes involved in the biosynthesis of haem d(1) in two heliobacterial species, Heliobacillus mobilis and Heliophilum fasciatum. The gene organization of the cluster is conserved between the two species, and contains a complete set of genes that lead to the biosynthesis of uroporphyrinogen III and genes thought to be involved in the late steps of haem d(1) biosynthesis. Detailed bioinformatics analysis of some of the proteins encoded in the gene cluster revealed important clues to the precise biochemical roles of the proteins in the biosynthesis of haem d(1), as well as the membrane transport and insertion of haem d(1) into an apocytochrome during the maturation of cytochrome cd(1).
Figures





Similar articles
-
Leptospira spp. possess a complete haem biosynthetic pathway and are able to use exogenous haem sources.Mol Microbiol. 2003 Aug;49(3):745-54. doi: 10.1046/j.1365-2958.2003.03589.x. Mol Microbiol. 2003. PMID: 12864856
-
Sequence of the core antenna domain from the anoxygenic phototroph Heliophilum fasciatum: implications for diversity of reaction center type I.Curr Microbiol. 2004 Jun;48(6):438-40. doi: 10.1007/s00284-003-4221-3. Curr Microbiol. 2004. PMID: 15170240
-
d(1) haem biogenesis - assessing the roles of three nir gene products.FEBS J. 2009 Nov;276(21):6399-411. doi: 10.1111/j.1742-4658.2009.07354.x. Epub 2009 Oct 1. FEBS J. 2009. PMID: 19796169
-
Sulfate-reducing bacteria reveal a new branch of tetrapyrrole metabolism.Adv Microb Physiol. 2012;61:267-95. doi: 10.1016/B978-0-12-394423-8.00007-X. Adv Microb Physiol. 2012. PMID: 23046956 Review.
-
Biochemistry, regulation and genomics of haem biosynthesis in prokaryotes.Adv Microb Physiol. 2002;46:257-318. doi: 10.1016/s0065-2911(02)46006-7. Adv Microb Physiol. 2002. PMID: 12073655 Review.
Cited by
-
Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin.Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2210-5. doi: 10.1073/pnas.1416285112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646457 Free PMC article.
-
Comparative Genomics of the Genus Porphyromonas Identifies Adaptations for Heme Synthesis within the Prevalent Canine Oral Species Porphyromonas cangingivalis.Genome Biol Evol. 2015 Nov 13;7(12):3397-413. doi: 10.1093/gbe/evv220. Genome Biol Evol. 2015. PMID: 26568374 Free PMC article.
-
Phenotypes and gene expression profiles of Saccharopolyspora erythraea rifampicin-resistant (rif) mutants affected in erythromycin production.Microb Cell Fact. 2009 Mar 30;8:18. doi: 10.1186/1475-2859-8-18. Microb Cell Fact. 2009. PMID: 19331655 Free PMC article.
-
Evolutionary relationships between heme-binding ferredoxin α + β barrels.BMC Bioinformatics. 2016 Apr 18;17:168. doi: 10.1186/s12859-016-1033-6. BMC Bioinformatics. 2016. PMID: 27089923 Free PMC article.
-
The structure, function and properties of sirohaem decarboxylase--an enzyme with structural homology to a transcription factor family that is part of the alternative haem biosynthesis pathway.Mol Microbiol. 2014 Jul;93(2):247-61. doi: 10.1111/mmi.12656. Epub 2014 Jun 18. Mol Microbiol. 2014. PMID: 24865947 Free PMC article.
References
-
- Azuaje F, Al-Shahrour F, Dopazo J. Ontology-driven approaches to analyzing data in functional genomics. Methods Mol Biol. 2006;316:67–86. - PubMed
-
- Beale SI. Biosynthesis and structures of porphyrins and hemes. In: Blankenship RE, Madigan MT, Bauer CE, editors. Anoxygenic Photosynthetic Bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 153–177.
-
- Beale SI. Tetrapyrrole biosynthesis in bacteria. In: Lederberg J, editor. Encyclopedia of Microbiology. 2. Vol. 4. San Diego, CA: Academic Press; 2000. pp. 558–570.
-
- Beer-Romero P, Gest H. Heliobacillus mobilis, a peritrichously flagellated anoxyphototroph containing bacteriochlorophyll g. FEMS Microbiol Lett. 1987;41:109–114.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

