Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody
- PMID: 17907804
- PMCID: PMC2323298
- DOI: 10.1371/journal.ppat.0030138
Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody
Erratum in
- PLoS Pathog. 2007 Nov;3(11):e172
Abstract
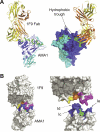
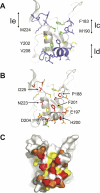
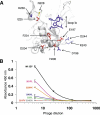
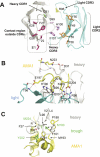
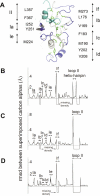
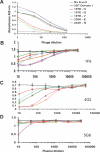
Identifying functionally critical regions of the malaria antigen AMA1 (apical membrane antigen 1) is necessary to understand the significance of the polymorphisms within this antigen for vaccine development. The crystal structure of AMA1 in complex with the Fab fragment of inhibitory monoclonal antibody 1F9 reveals that 1F9 binds to the AMA1 solvent-exposed hydrophobic trough, confirming its importance. 1F9 uses the heavy and light chain complementarity-determining regions (CDRs) to wrap around the polymorphic loops adjacent to the trough, but uses a ridge of framework residues to bind to the hydrophobic trough. The resulting 1F9-AMA1-combined buried surface of 2,470 A(2) is considerably larger than previously reported Fab-antigen interfaces. Mutations of polymorphic AMA1 residues within the 1F9 epitope disrupt 1F9 binding and dramatically reduce the binding of affinity-purified human antibodies. Moreover, 1F9 binding to AMA1 is competed by naturally acquired human antibodies, confirming that the 1F9 epitope is a frequent target of immunological attack.
Conflict of interest statement
Figures
References
-
- Breman JG, Alilio MS, Mills A. Conquering the intolerable burden of malaria: What's new, what's needed: a summary. Am J Trop Med Hyg. 2004;71:1–15. - PubMed
-
- Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–685. - PubMed
-
- Baird JK. Host age as a determinant of naturally acquired immunity to Plasmodium falciparum. Parasitol Today. 1995;11:105–111. - PubMed
-
- Cohen S, McGregor IA, Carrington SC. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. - PubMed
-
- Cohen S, Butcher GA. Properties of protective malarial antibody. Nature. 1970;225:732–734. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

