Gene expression programs of human smooth muscle cells: tissue-specific differentiation and prognostic significance in breast cancers
- PMID: 17907811
- PMCID: PMC1994710
- DOI: 10.1371/journal.pgen.0030164
Gene expression programs of human smooth muscle cells: tissue-specific differentiation and prognostic significance in breast cancers
Abstract
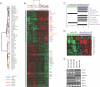
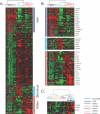
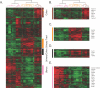
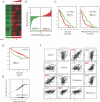
Smooth muscle is present in a wide variety of anatomical locations, such as blood vessels, various visceral organs, and hair follicles. Contraction of smooth muscle is central to functions as diverse as peristalsis, urination, respiration, and the maintenance of vascular tone. Despite the varied physiological roles of smooth muscle cells (SMCs), we possess only a limited knowledge of the heterogeneity underlying their functional and anatomic specializations. As a step toward understanding the intrinsic differences between SMCs from different anatomical locations, we used DNA microarrays to profile global gene expression patterns in 36 SMC samples from various tissues after propagation under defined conditions in cell culture. Significant variations were found between the cells isolated from blood vessels, bronchi, and visceral organs. Furthermore, pervasive differences were noted within the visceral organ subgroups that appear to reflect the distinct molecular pathways essential for organogenesis as well as those involved in organ-specific contractile and physiological properties. Finally, we sought to understand how this diversity may contribute to SMC-involving pathology. We found that a gene expression signature of the responses of vascular SMCs to serum exposure is associated with a significantly poorer prognosis in human cancers, potentially linking vascular injury response to tumor progression.
Conflict of interest statement
Competing interests. POB is a cofounder of the Public Library of Science and is on its board of directors.
Figures

References
-
- Owens GK, Wise G. Regulation of differentiation/maturation in vascular smooth muscle cells by hormones and growth factors. Agents Actions Suppl. 1997;48:3–24. - PubMed
-
- Kumar MS, Owens GK. Combinatorial control of smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2003;23:737–747. - PubMed
-
- Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. - PubMed
-
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

