Metal ion-dependent, reversible, protein filament formation by designed beta-roll polypeptides
- PMID: 17908326
- PMCID: PMC2174480
- DOI: 10.1186/1472-6807-7-63
Metal ion-dependent, reversible, protein filament formation by designed beta-roll polypeptides
Abstract
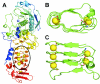
Background: A right-handed, calcium-dependent beta-roll structure found in secreted proteases and repeat-in-toxin proteins was used as a template for the design of minimal, soluble, monomeric polypeptides that would fold in the presence of Ca2+. Two polypeptides were synthesised to contain two and four metal-binding sites, respectively, and exploit stacked tryptophan pairs to stabilise the fold and report on the conformational state of the polypeptide.
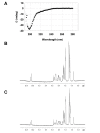
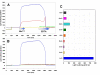
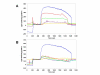
Results: Initial analysis of the two polypeptides in the presence of calcium suggested the polypeptides were disordered. The addition of lanthanum to these peptides caused aggregation. Upon further study by right angle light scattering and electron microscopy, the aggregates were identified as ordered protein filaments that required lanthanum to polymerize. These filaments could be disassembled by the addition of a chelating agent. A simple head-to-tail model is proposed for filament formation that explains the metal ion-dependency. The model is supported by the capping of one of the polypeptides with biotin, which disrupts filament formation and provides the ability to control the average length of the filaments.
Conclusion: Metal ion-dependent, reversible protein filament formation is demonstrated for two designed polypeptides. The polypeptides form filaments that are approximately 3 nm in diameter and several hundred nm in length. They are not amyloid-like in nature as demonstrated by their behaviour in the presence of congo red and thioflavin T. A capping strategy allows for the control of filament length and for potential applications including the "decoration" of a protein filament with various functional moieties.
Figures




Similar articles
-
Structure-based design and study of non-amyloidogenic, double N-methylated IAPP amyloid core sequences as inhibitors of IAPP amyloid formation and cytotoxicity.J Mol Biol. 2002 Jan 18;315(3):339-50. doi: 10.1006/jmbi.2001.5244. J Mol Biol. 2002. PMID: 11786016
-
Mechanism of formation of amyloid protofibrils of barstar from soluble oligomers: evidence for multiple steps and lateral association coupled to conformational conversion.J Mol Biol. 2007 Apr 6;367(4):1186-204. doi: 10.1016/j.jmb.2007.01.039. Epub 2007 Jan 20. J Mol Biol. 2007. PMID: 17292913
-
Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. An experimental support for the key role of the phenylalanine residue in amyloid formation.J Biol Chem. 2001 Sep 7;276(36):34156-61. doi: 10.1074/jbc.M102883200. Epub 2001 Jul 9. J Biol Chem. 2001. PMID: 11445568
-
Molecular structure of amyloid fibrils: insights from solid-state NMR.Q Rev Biophys. 2006 Feb;39(1):1-55. doi: 10.1017/S0033583506004173. Epub 2006 Jun 13. Q Rev Biophys. 2006. PMID: 16772049 Review.
-
Amyloid peptide pores and the beta sheet conformation.Adv Exp Med Biol. 2010;677:150-67. doi: 10.1007/978-1-4419-6327-7_13. Adv Exp Med Biol. 2010. PMID: 20687488 Review.
Cited by
-
Block V RTX Domain of Adenylate Cyclase from Bordetella pertussis: A Conformationally Dynamic Scaffold for Protein Engineering Applications.Toxins (Basel). 2017 Sep 17;9(9):289. doi: 10.3390/toxins9090289. Toxins (Basel). 2017. PMID: 28926974 Free PMC article. Review.
-
Nanometer to millimeter scale peptide-porphyrin materials.Biomacromolecules. 2010 Oct 11;11(10):2602-9. doi: 10.1021/bm100540t. Biomacromolecules. 2010. PMID: 20804210 Free PMC article.
-
Zinc(II) modulates specifically amyloid formation and structure in model peptides.J Biol Inorg Chem. 2011 Feb;16(2):333-40. doi: 10.1007/s00775-010-0729-8. J Biol Inorg Chem. 2011. PMID: 21061029
-
Evaluation of a symmetry-based strategy for assembling protein complexes.RSC Adv. 2011 Oct 21;1(6):1004-1012. doi: 10.1039/C1RA00282A. RSC Adv. 2011. PMID: 23293744 Free PMC article.
-
Metal-dependent assembly of a protein nano-cage.Protein Sci. 2019 Sep;28(9):1620-1629. doi: 10.1002/pro.3676. Epub 2019 Aug 6. Protein Sci. 2019. PMID: 31278804 Free PMC article.
References
-
- Sen A, Baxa U, Simon MN, Wall JS, Sabate R, Saupe SJ, Steven AC. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

