Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets
- PMID: 17908789
- PMCID: PMC2169186
- DOI: 10.1128/MCB.01493-07
Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets
Abstract
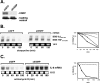
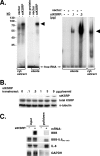
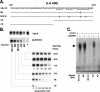
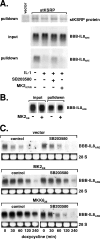
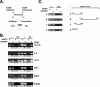
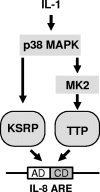
mRNA stability is a major determinant of inflammatory gene expression. Rapid degradation of interleukin-8 (IL-8) mRNA is imposed by a bipartite AU-rich element (ARE) in the 3' untranslated region (R. Winzen et al., Mol. Cell. Biol. 24:4835-4847, 2004). Small interfering RNA-mediated knockdown of the ARE-binding protein KSRP resulted in stabilization of IL-8 mRNA or of a beta-globin reporter mRNA containing the IL-8 ARE. Rapid deadenylation was impaired, indicating a crucial role for KSRP in this step of mRNA degradation. The two IL-8 ARE domains both contribute to interaction with KSRP, corresponding to the importance of both domains for rapid degradation. Exposure to the inflammatory cytokine IL-1 has been shown to stabilize IL-8 mRNA through p38 mitogen-activated protein (MAP) kinase and MK2. IL-1 treatment impaired the interaction of KSRP with the IL-8 ARE in a manner dependent on p38 MAP kinase but apparently independent of MK2. Instead, evidence that TTP, a target of MK2, can also destabilize the IL-8 ARE reporter mRNA is presented. In a comprehensive approach to identify mRNAs controlled by KSRP, two criteria were evaluated by microarray analysis of (i) association of mRNAs with KSRP in pulldown assays and (ii) increased amounts in KSRP knockdown cells. According to both criteria, a group of 100 mRNAs is controlled by KSRP, many of which are unstable and encode proteins involved in inflammation. These results indicate that KSRP functions as a limiting factor in inflammatory gene expression.
Figures


References
-
- Ben-Levy, R., S. Hooper, R. Wilson, H. F. Paterson, and C. J. Marshall. 1998. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8:1049-1057. - PubMed
-
- Briata, P., S. V. Forcales, M. Ponassi, G. Corte, C. Y. Chen, M. Karin, P. L. Puri, and R. Gherzi. 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20:891-903. - PubMed
-
- Brook, M., C. R. Tchen, T. Santalucia, J. McIlrath, J. S. Arthur, J. Saklatvala, and A. R. Clark. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26:2408-2418. - PMC - PubMed
-
- Brooks, S. A., J. E. Connolly, and W. F. Rigby. 2004. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 172:7263-7271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
