Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis
- PMID: 17908800
- PMCID: PMC2169415
- DOI: 10.1128/MCB.00821-07
Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis
Abstract
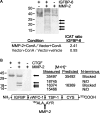
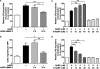
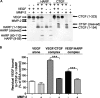
Matrix metalloproteinases (MMPs) exert both pro- and antiangiogenic functions by the release of cytokines or proteolytically generated angiogenic inhibitors from extracellular matrix and basement membrane remodeling. In the Mmp2-/- mouse neovascularization is greatly reduced, but the mechanistic aspects of this remain unclear. Using isotope-coded affinity tag labeling of proteins analyzed by multidimensional liquid chromatography and tandem mass spectrometry we explored proteome differences between Mmp2-/- cells and those rescued by MMP-2 transfection. Proteome signatures that are hallmarks of proteolysis revealed cleavage of many known MMP-2 substrates in the cellular context. Proteomic evidence of MMP-2 processing of novel substrates was found. Insulin-like growth factor binding protein 6, follistatin-like 1, and cystatin C protein cleavage by MMP-2 was biochemically confirmed, and the cleavage sites in heparin affin regulatory peptide (HARP; pleiotrophin) and connective tissue growth factor (CTGF) were sequenced by matrix-assisted laser desorption ionization-time of flight mass spectrometry. MMP-2 processing of HARP and CTGF released vascular endothelial growth factor (VEGF) from angiogenic inhibitory complexes. The cleaved HARP N-terminal domain increased HARP-induced cell proliferation, whereas the HARP C-terminal domain was antagonistic and decreased cell proliferation and migration. Hence the unmasking of cytokines, such as VEGF, by metalloproteinase processing of their binding proteins is a new mechanism in the control of cytokine activation and angiogenesis.
Figures


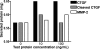

Similar articles
-
Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165.J Biol Chem. 2002 Sep 27;277(39):36288-95. doi: 10.1074/jbc.M201674200. Epub 2002 Jul 11. J Biol Chem. 2002. PMID: 12114504
-
Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics.Mol Cell Proteomics. 2010 May;9(5):894-911. doi: 10.1074/mcp.M000050-MCP201. Epub 2010 Mar 20. Mol Cell Proteomics. 2010. PMID: 20305284 Free PMC article.
-
Heparin affin regulatory peptide binds to vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis.Oncogene. 2004 Mar 4;23(9):1745-53. doi: 10.1038/sj.onc.1206879. Oncogene. 2004. PMID: 15001987
-
Proteomic validation of protease drug targets: pharmacoproteomics of matrix metalloproteinase inhibitor drugs using isotope-coded affinity tag labelling and tandem mass spectrometry.Curr Pharm Des. 2007;13(3):263-70. doi: 10.2174/138161207779313524. Curr Pharm Des. 2007. PMID: 17313360 Review.
-
Regulation of matrix metalloproteinases 2 and 9 in corneal neovascularization.Chem Biol Drug Des. 2020 May;95(5):485-492. doi: 10.1111/cbdd.13529. Epub 2020 Feb 16. Chem Biol Drug Des. 2020. PMID: 31002472 Review.
Cited by
-
The metastasis inducer CCN1 (CYR61) activates the fatty acid synthase (FASN)-driven lipogenic phenotype in breast cancer cells.Oncoscience. 2016 Jul 22;3(7-8):242-257. doi: 10.18632/oncoscience.314. eCollection 2016. Oncoscience. 2016. PMID: 27713913 Free PMC article.
-
Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: dynamics of membrane type 1 matrix metalloproteinase-mediated membrane protein shedding.Mol Cell Biol. 2008 Aug;28(15):4896-914. doi: 10.1128/MCB.01775-07. Epub 2008 May 27. Mol Cell Biol. 2008. PMID: 18505826 Free PMC article.
-
Skeletal overexpression of connective tissue growth factor impairs bone formation and causes osteopenia.Endocrinology. 2008 Sep;149(9):4374-81. doi: 10.1210/en.2008-0254. Epub 2008 Jun 5. Endocrinology. 2008. PMID: 18535099 Free PMC article.
-
A Cystatin C Cleavage ELISA Assay as a Quality Control Tool for Determining Sub-Optimal Storage Conditions of Cerebrospinal Fluid Samples in Alzheimer's Disease Research.J Alzheimers Dis. 2021;83(3):1367-1377. doi: 10.3233/JAD-210741. J Alzheimers Dis. 2021. PMID: 34420976 Free PMC article.
-
Proteolytic post-translational modification of proteins: proteomic tools and methodology.Mol Cell Proteomics. 2013 Dec;12(12):3532-42. doi: 10.1074/mcp.M113.031310. Epub 2013 Jul 25. Mol Cell Proteomics. 2013. PMID: 23887885 Free PMC article. Review.
References
-
- Amthor, H., G. Nicholas, I. McKinnell, C. F. Kemp, M. Sharma, R. Kambadur, and K. Patel. 2004. Follistatin complexes myostatin and antagonises myostatin-mediated inhibition of myogenesis. Dev. Biol. 270:19-30. - PubMed
-
- Anand-Apte, B., M. S. Pepper, E. Voest, R. Montesano, B. Olsen, G. Murphy, S. S. Apte, and B. Zetter. 1997. Inhibition of angiogenesis by tissue inhibitor of metalloproteinase-3. Investig. Ophthalmol. Vis. Sci. 38:817-823. - PubMed
-
- Bach, L. A. 2005. IGFBP-6 five years on; not so ‘forgotten’? Growth Horm. IGF Res. 15:185-192. - PubMed
-
- Balbin, M., A. Fueyo, A. M. Tester, A. M. Pendas, A. S. Pitiot, A. Astudillo, C. M. Overall, S. D. Shapiro, and C. Lopez-Otín. 2003. Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nat. Genet. 35:252-257. - PubMed
-
- Ball, D. K., A. W. Rachfal, S. A. Kemper, and D. R. Brigstock. 2003. The heparin-binding 10 kDa fragment of connective tissue growth factor (CTGF) containing module 4 alone stimulates cell adhesion. J. Endocrinol. 176:R1-R7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
