The nature of telomere fusion and a definition of the critical telomere length in human cells
- PMID: 17908935
- PMCID: PMC1993879
- DOI: 10.1101/gad.439107
The nature of telomere fusion and a definition of the critical telomere length in human cells
Abstract
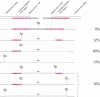
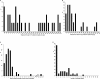
The loss of telomere function can result in telomeric fusion events that lead to the types of genomic rearrangements, such as nonreciprocal translocations, that typify early-stage carcinogenesis. By using single-molecule approaches to characterize fusion events, we provide a functional definition of fusogenic telomeres in human cells. We show that approximately half of the fusion events contained no canonical telomere repeats at the fusion point; of those that did, the longest was 12.8 repeats. Furthermore, in addition to end-replication losses, human telomeres are subjected to large-scale deletion events that occur in the presence or absence of telomerase. Here we show that these telomeres are fusogenic, and thus despite the majority of telomeres being maintained at a stable length in normal human cells, a subset of stochastically shortened telomeres can potentially cause chromosomal instability. Telomere fusion was accompanied by the deletion of one or both telomeres extending several kilobases into the telomere-adjacent DNA, and microhomology was observed at the fusion points. This contrasted with telomere fusion that was observed following the experimental disruption of TRF2. The distinct error-prone mutational profile of fusion between critically shortened telomeres in human cells was reminiscent of Ku-independent microhomology-mediated end-joining.
Figures




References
-
- Artandi S.E., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Gottlieb G.J., Chin L., DePinho R.A., Chin L., DePinho R.A., DePinho R.A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. - PubMed
-
- Atkin N.B. Lack of reciprocal translocations in carcinomas. Cancer Genet. Cytogenet. 1986;21:275–278. - PubMed
-
- Auersperg N. Long-term cultivation of hypodiploid human tumor cells. J. Natl. Cancer Inst. 1964;32:135–163. - PubMed
-
- Bailey S.M., Cornforth M.N., Kurimasa A., Chen D.J., Goodwin E.H., Cornforth M.N., Kurimasa A., Chen D.J., Goodwin E.H., Kurimasa A., Chen D.J., Goodwin E.H., Chen D.J., Goodwin E.H., Goodwin E.H. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
