Structure of Alzheimer's disease amyloid precursor protein copper-binding domain at atomic resolution
- PMID: 17909280
- PMCID: PMC2339725
- DOI: 10.1107/S1744309107041139
Structure of Alzheimer's disease amyloid precursor protein copper-binding domain at atomic resolution
Abstract
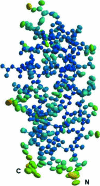
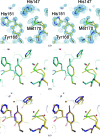
Amyloid precursor protein (APP) plays a central role in the pathogenesis of Alzheimer's disease, as its cleavage generates the Abeta peptide that is toxic to cells. APP is able to bind Cu2+ and reduce it to Cu+ through its copper-binding domain (CuBD). The interaction between Cu2+ and APP leads to a decrease in Abeta production and to alleviation of the symptoms of the disease in mouse models. Structural studies of CuBD have been undertaken in order to better understand the mechanism behind the process. Here, the crystal structure of CuBD in the metal-free form determined to ultrahigh resolution (0.85 A) is reported. The structure shows that the copper-binding residues of CuBD are rather rigid but that Met170, which is thought to be the electron source for Cu2+ reduction, adopts two different side-chain conformations. These observations shed light on the copper-binding and redox mechanisms of CuBD. The structure of CuBD at atomic resolution provides an accurate framework for structure-based design of molecules that will deplete Abeta production.
Figures
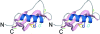
Similar articles
-
Crystallization and preliminary crystallographic studies of the copper-binding domain of the amyloid precursor protein of Alzheimer's disease.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Jan 1;61(Pt 1):93-5. doi: 10.1107/S1744309104029744. Epub 2004 Dec 2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005. PMID: 16508101 Free PMC article.
-
Structural studies of the Alzheimer's amyloid precursor protein copper-binding domain reveal how it binds copper ions.J Mol Biol. 2007 Mar 16;367(1):148-61. doi: 10.1016/j.jmb.2006.12.041. Epub 2006 Dec 21. J Mol Biol. 2007. PMID: 17239395
-
Contrasting, species-dependent modulation of copper-mediated neurotoxicity by the Alzheimer's disease amyloid precursor protein.J Neurosci. 2002 Jan 15;22(2):365-76. doi: 10.1523/JNEUROSCI.22-02-00365.2002. J Neurosci. 2002. PMID: 11784781 Free PMC article.
-
Copper binding to the Alzheimer's disease amyloid precursor protein.Eur Biophys J. 2008 Mar;37(3):269-79. doi: 10.1007/s00249-007-0234-3. Epub 2007 Nov 21. Eur Biophys J. 2008. PMID: 18030462 Free PMC article. Review.
-
Copper brain homeostasis: role of amyloid precursor protein and prion protein.IUBMB Life. 2005 Sep;57(9):645-50. doi: 10.1080/15216540500264620. IUBMB Life. 2005. PMID: 16203684 Review.
Cited by
-
Copper in the brain and Alzheimer's disease.J Biol Inorg Chem. 2010 Jan;15(1):61-76. doi: 10.1007/s00775-009-0600-y. Epub 2009 Oct 28. J Biol Inorg Chem. 2010. PMID: 19862561 Review.
-
Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain.J Neurosci. 2014 Aug 13;34(33):11159-72. doi: 10.1523/JNEUROSCI.0180-14.2014. J Neurosci. 2014. PMID: 25122912 Free PMC article.
-
Alzheimer's disease--a panorama glimpse.Int J Mol Sci. 2014 Jul 16;15(7):12631-50. doi: 10.3390/ijms150712631. Int J Mol Sci. 2014. PMID: 25032844 Free PMC article. Review.
-
Structural biology of cell surface receptors implicated in Alzheimer's disease.Biophys Rev. 2021 Nov 18;14(1):233-255. doi: 10.1007/s12551-021-00903-9. eCollection 2022 Feb. Biophys Rev. 2021. PMID: 35340615 Free PMC article. Review.
-
Insights into the distribution and abundance of the ubiquitous candidatus Saccharibacteria phylum following tag pyrosequencing.Sci Rep. 2014 Feb 4;4:3957. doi: 10.1038/srep03957. Sci Rep. 2014. PMID: 24492458 Free PMC article.
References
-
- Barnham, K. J., McKinstry, W. J., Multhaup, G., Galatis, D., Morton, C. J., Curtain, C. C., Williamson, N. A., White, A. R., Hinds, M. G., Norton, R. S., Beyreuther, K., Masters, C. L., Parker, M. W. & Cappai, R. (2003). J. Biol. Chem.278, 17401–17407. - PubMed
-
- Borchardt, T., Schmidt, C., Camakaris, J., Cappai, R., Masters, C. L., Beyreuther, K. & Multhaup, G. (2000). Cell. Mol. Biol.46, 785–795. - PubMed
-
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

