BNDB - the Biochemical Network Database
- PMID: 17910766
- PMCID: PMC2092437
- DOI: 10.1186/1471-2105-8-367
BNDB - the Biochemical Network Database
Abstract
Background: Technological advances in high-throughput techniques and efficient data acquisition methods have resulted in a massive amount of life science data. The data is stored in numerous databases that have been established over the last decades and are essential resources for scientists nowadays. However, the diversity of the databases and the underlying data models make it difficult to combine this information for solving complex problems in systems biology. Currently, researchers typically have to browse several, often highly focused, databases to obtain the required information. Hence, there is a pressing need for more efficient systems for integrating, analyzing, and interpreting these data. The standardization and virtual consolidation of the databases is a major challenge resulting in a unified access to a variety of data sources.
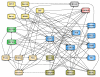
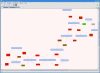
Description: We present the Biochemical Network Database (BNDB), a powerful relational database platform, allowing a complete semantic integration of an extensive collection of external databases. BNDB is built upon a comprehensive and extensible object model called BioCore, which is powerful enough to model most known biochemical processes and at the same time easily extensible to be adapted to new biological concepts. Besides a web interface for the search and curation of the data, a Java-based viewer (BiNA) provides a powerful platform-independent visualization and navigation of the data. BiNA uses sophisticated graph layout algorithms for an interactive visualization and navigation of BNDB.
Conclusion: BNDB allows a simple, unified access to a variety of external data sources. Its tight integration with the biochemical network library BN++ offers the possibility for import, integration, analysis, and visualization of the data. BNDB is freely accessible at http://www.bndb.org.
Figures




Similar articles
-
Browsing multidimensional molecular networks with the generic network browser (N-Browse).Curr Protoc Bioinformatics. 2008 Sep;Chapter 9:9.11.1-9.11.21. doi: 10.1002/0471250953.bi0911s23. Curr Protoc Bioinformatics. 2008. PMID: 18819079 Free PMC article.
-
A database and tool, IM Browser, for exploring and integrating emerging gene and protein interaction data for Drosophila.BMC Bioinformatics. 2006 Apr 7;7:195. doi: 10.1186/1471-2105-7-195. BMC Bioinformatics. 2006. PMID: 16603075 Free PMC article.
-
PATIKAweb: a Web interface for analyzing biological pathways through advanced querying and visualization.Bioinformatics. 2006 Feb 1;22(3):374-5. doi: 10.1093/bioinformatics/bti776. Epub 2005 Nov 15. Bioinformatics. 2006. PMID: 16287939
-
Searching the MINT database for protein interaction information.Curr Protoc Bioinformatics. 2008 Jun;Chapter 8:8.5.1-8.5.13. doi: 10.1002/0471250953.bi0805s22. Curr Protoc Bioinformatics. 2008. PMID: 18551417 Review.
-
Towards Semantic e-Science for Traditional Chinese Medicine.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S6. doi: 10.1186/1471-2105-8-S3-S6. BMC Bioinformatics. 2007. PMID: 17493289 Free PMC article. Review.
Cited by
-
BiNA: a visual analytics tool for biological network data.PLoS One. 2014 Feb 13;9(2):e87397. doi: 10.1371/journal.pone.0087397. eCollection 2014. PLoS One. 2014. PMID: 24551056 Free PMC article.
-
SBMLsqueezer: a CellDesigner plug-in to generate kinetic rate equations for biochemical networks.BMC Syst Biol. 2008 Apr 30;2:39. doi: 10.1186/1752-0509-2-39. BMC Syst Biol. 2008. PMID: 18447902 Free PMC article.
-
VANTED v2: a framework for systems biology applications.BMC Syst Biol. 2012 Nov 10;6:139. doi: 10.1186/1752-0509-6-139. BMC Syst Biol. 2012. PMID: 23140568 Free PMC article.
-
GeneTrailExpress: a web-based pipeline for the statistical evaluation of microarray experiments.BMC Bioinformatics. 2008 Dec 22;9:552. doi: 10.1186/1471-2105-9-552. BMC Bioinformatics. 2008. PMID: 19099609 Free PMC article.
-
Semantic integration of data on transcriptional regulation.Bioinformatics. 2010 Jul 1;26(13):1651-61. doi: 10.1093/bioinformatics/btq231. Epub 2010 Apr 28. Bioinformatics. 2010. PMID: 20427517 Free PMC article.
References
-
- Cassman M, Arkin A, Doyle F, Katagiri F, Lauffenburger DA, Stokes C. Tech rep. World Technology Evaluation Center (WTEC); 2005. Assessment of International Research and Development in Systems Biology.
-
- Hernandez T, Kambhampati S. Integration of Biological Sources: Current Systems and Challenges Ahead. SIGMOD Rec. 2004;33:51–60. doi: 10.1145/1031570.1031583. - DOI
-
- Etzold T, Argos P. SRS – an indexing and retrieval tool for flat file data libraries. Comput Appl Biosci. 1993;9:49–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

