SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins
- PMID: 17912346
- PMCID: PMC1989135
- DOI: 10.1371/journal.pone.0000967
SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins
Abstract
Background: Short linear motifs (SLiMs) in proteins are functional microdomains of fundamental importance in many biological systems. SLiMs typically consist of a 3 to 10 amino acid stretch of the primary protein sequence, of which as few as two sites may be important for activity, making identification of novel SLiMs extremely difficult. In particular, it can be very difficult to distinguish a randomly recurring "motif" from a truly over-represented one. Incorporating ambiguous amino acid positions and/or variable-length wildcard spacers between defined residues further complicates the matter.
Methodology/principal findings: In this paper we present two algorithms. SLiMBuild identifies convergently evolved, short motifs in a dataset of proteins. Motifs are built by combining dimers into longer patterns, retaining only those motifs occurring in a sufficient number of unrelated proteins. Motifs with fixed amino acid positions are identified and then combined to incorporate amino acid ambiguity and variable-length wildcard spacers. The algorithm is computationally efficient compared to alternatives, particularly when datasets include homologous proteins, and provides great flexibility in the nature of motifs returned. The SLiMChance algorithm estimates the probability of returned motifs arising by chance, correcting for the size and composition of the dataset, and assigns a significance value to each motif. These algorithms are implemented in a software package, SLiMFinder. SLiMFinder default settings identify known SLiMs with 100% specificity, and have a low false discovery rate on random test data.
Conclusions/significance: The efficiency of SLiMBuild and low false discovery rate of SLiMChance make SLiMFinder highly suited to high throughput motif discovery and individual high quality analyses alike. Examples of such analyses on real biological data, and how SLiMFinder results can help direct future discoveries, are provided. SLiMFinder is freely available for download under a GNU license from http://bioinformatics.ucd.ie/shields/software/slimfinder/.
Conflict of interest statement
Figures

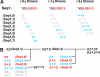


Similar articles
-
SLiMFinder: a web server to find novel, significantly over-represented, short protein motifs.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W534-9. doi: 10.1093/nar/gkq440. Epub 2010 May 23. Nucleic Acids Res. 2010. PMID: 20497999 Free PMC article.
-
Estimation and efficient computation of the true probability of recurrence of short linear protein sequence motifs in unrelated proteins.BMC Bioinformatics. 2010 Jan 7;11:14. doi: 10.1186/1471-2105-11-14. BMC Bioinformatics. 2010. PMID: 20055997 Free PMC article.
-
Masking residues using context-specific evolutionary conservation significantly improves short linear motif discovery.Bioinformatics. 2009 Feb 15;25(4):443-50. doi: 10.1093/bioinformatics/btn664. Epub 2009 Jan 9. Bioinformatics. 2009. PMID: 19136552
-
Bioinformatics Approaches for Predicting Disordered Protein Motifs.Adv Exp Med Biol. 2015;870:291-318. doi: 10.1007/978-3-319-20164-1_9. Adv Exp Med Biol. 2015. PMID: 26387106 Review.
-
Computational prediction of short linear motifs from protein sequences.Methods Mol Biol. 2015;1268:89-141. doi: 10.1007/978-1-4939-2285-7_6. Methods Mol Biol. 2015. PMID: 25555723 Review.
Cited by
-
A novel binding site on the cryptic intervening domain is a motif-dependent regulator of O-GlcNAc transferase.Res Sq [Preprint]. 2023 Feb 2:rs.3.rs-2531412. doi: 10.21203/rs.3.rs-2531412/v1. Res Sq. 2023. Update in: Nat Chem Biol. 2023 Nov;19(11):1423-1431. doi: 10.1038/s41589-023-01422-2. PMID: 36778302 Free PMC article. Updated. Preprint.
-
Predicting binding within disordered protein regions to structurally characterised peptide-binding domains.PLoS One. 2013 Sep 3;8(9):e72838. doi: 10.1371/journal.pone.0072838. eCollection 2013. PLoS One. 2013. PMID: 24019881 Free PMC article.
-
PMS: a panoptic motif search tool.PLoS One. 2013 Dec 4;8(12):e80660. doi: 10.1371/journal.pone.0080660. eCollection 2013. PLoS One. 2013. PMID: 24324619 Free PMC article.
-
Advanced computational approaches to understand protein aggregation.Biophys Rev (Melville). 2024 Apr 24;5(2):021302. doi: 10.1063/5.0180691. eCollection 2024 Jun. Biophys Rev (Melville). 2024. PMID: 38681860 Free PMC article. Review.
-
Evaluating caveolin interactions: do proteins interact with the caveolin scaffolding domain through a widespread aromatic residue-rich motif?PLoS One. 2012;7(9):e44879. doi: 10.1371/journal.pone.0044879. Epub 2012 Sep 17. PLoS One. 2012. PMID: 23028656 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

