Design and pre-clinical evaluation of a universal HIV-1 vaccine
- PMID: 17912361
- PMCID: PMC1991584
- DOI: 10.1371/journal.pone.0000984
Design and pre-clinical evaluation of a universal HIV-1 vaccine
Erratum in
- PLoS One. 2011;6(3). doi: 10.1371/annotation/fca26a4f-42c1-4772-a19e-aa9d96c4eeb2
Abstract
Background: One of the big roadblocks in development of HIV-1/AIDS vaccines is the enormous diversity of HIV-1, which could limit the value of any HIV-1 vaccine candidate currently under test.
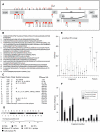
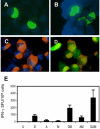
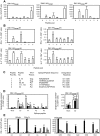
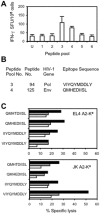
Methodology and findings: To address the HIV-1 variation, we designed a novel T cell immunogen, designated HIV(CONSV), by assembling the 14 most conserved regions of the HIV-1 proteome into one chimaeric protein. Each segment is a consensus sequence from one of the four major HIV-1 clades A, B, C and D, which alternate to ensure equal clade coverage. The gene coding for the HIV(CONSV) protein was inserted into the three most studied vaccine vectors, plasmid DNA, human adenovirus serotype 5 and modified vaccine virus Ankara (MVA), and induced HIV-1-specific T cell responses in mice. We also demonstrated that these conserved regions prime CD8(+) and CD4(+) T cell to highly conserved epitopes in humans and that these epitopes, although usually subdominant, generate memory T cells in patients during natural HIV-1 infection.
Significance: Therefore, this vaccine approach provides an attractive and testable alternative for overcoming the HIV-1 variability, while focusing T cell responses on regions of the virus that are less likely to mutate and escape. Furthermore, this approach has merit in the simplicity of design and delivery, requiring only a single immunogen to provide extensive coverage of global HIV-1 population diversity.
Conflict of interest statement
Figures
References
-
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. - PubMed
-
- McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. - PubMed
-
- Duerr A, Wasserheit JN, Corey L. HIV vaccines: new frontiers in vaccine development. Clin Infect Dis. 2006;43:500–511. - PubMed
-
- Hanke T, McMichael AJ, Dorrell L. Clinical experience with plasmid DNA- and modified vaccinia vaccine Ankara (MVA)-vectored HIV-1 clade A vaccine inducing T cells. J Gen Virol. 2007;88:1–12. - PubMed
-
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

