We gather together: insulators and genome organization
- PMID: 17913488
- PMCID: PMC2215060
- DOI: 10.1016/j.gde.2007.08.005
We gather together: insulators and genome organization
Abstract
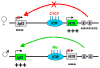
When placed between an enhancer and promoter, certain DNA sequence elements inhibit enhancer-stimulated gene expression. The best characterized of these enhancer-blocking insulators, gypsy in Drosophila and the CTCF-binding element in vertebrates and flies, stabilize contacts between distant genomic regulatory sites leading to the formation of loop domains. Current results show that CTCF mediates long-range contacts in the mouse beta-globin locus and at the Igf2/H19-imprinted locus. Recently described active chromatin hubs and transcription factories also involve long-range interactions; it is likely that CTCF interferes with their formation when acting as an insulator. The properties of CTCF, and its newly described genomic distribution, suggest that it may play an important role in large-scale nuclear architecture, perhaps mediated by the co-factors with which it interacts in vivo.
Figures



References
-
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–116. - PubMed
-
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. - PubMed
-
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
-
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

