The Ser176 of T4 endonuclease IV is crucial for the restricted and polarized dC-specific cleavage of single-stranded DNA implicated in restriction of dC-containing DNA in host Escherichia coli
- PMID: 17913749
- PMCID: PMC2175332
- DOI: 10.1093/nar/gkm722
The Ser176 of T4 endonuclease IV is crucial for the restricted and polarized dC-specific cleavage of single-stranded DNA implicated in restriction of dC-containing DNA in host Escherichia coli
Abstract
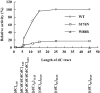
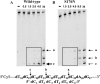
Endonuclease (Endo) IV encoded by denB of bacteriophage T4 is an enzyme that cleaves single-stranded (ss) DNA in a dC-specific manner. Also the growth of dC-substituted T4 phage and host Escherichia coli cells is inhibited by denB expression presumably because of the inhibitory effect on replication of dC-containing DNA. Recently, we have demonstrated that an efficient cleavage by Endo IV occurs exclusively at the 5'-proximal dC (dC1) within a hexameric or an extended sequence consisting of dC residues at the 5'-proximal and the 3'-proximal positions (dCs tract), in which a third dC residue within the tract affects the polarized cleavage and cleavage rate. Here we isolate and characterize two denB mutants, denB(W88R) and denB(S176N). Both mutant alleles have lost the detrimental effect on the host cell. Endo IV(W88R) shows no enzymatic activity (<0.4% of that of wild-type Endo IV). On the other hand, Endo IV(S176N) retains cleavage activity (17.5% of that of wild-type Endo IV), but has lost the polarized and restricted cleavage of a dCs tract, indicating that the Ser176 residue of Endo IV is implicated in the polarized cleavage of a dCs tract which brings about a detrimental effect on the replication of dC-containing DNA.
Figures
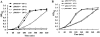


Similar articles
-
A hexanucleotide sequence (dC1-dC6 tract) restricts the dC-specific cleavage of single-stranded DNA by endonuclease IV of bacteriophage T4.Nucleic Acids Res. 2007;35(19):6681-9. doi: 10.1093/nar/gkm721. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17940096 Free PMC article.
-
Biochemical analysis of the substrate specificity and sequence preference of endonuclease IV from bacteriophage T4, a dC-specific endonuclease implicated in restriction of dC-substituted T4 DNA synthesis.Nucleic Acids Res. 2006;34(17):4743-51. doi: 10.1093/nar/gkl553. Epub 2006 Sep 13. Nucleic Acids Res. 2006. PMID: 16971463 Free PMC article.
-
Characterization of amino acid substitutions that severely alter the DNA repair functions of Escherichia coli endonuclease IV.Biochemistry. 1999 Mar 23;38(12):3615-23. doi: 10.1021/bi9824083. Biochemistry. 1999. PMID: 10090748
-
Use of an Escherichia coli mutant strain permits measurement of single-stranded apurinic-apyrimidinic endonuclease in crude extracts: studies with untransformed cells and cells transformed with plasmids containing the uvrC gene.J Bacteriol. 1983 Jun;154(3):1459-61. doi: 10.1128/jb.154.3.1459-1461.1983. J Bacteriol. 1983. PMID: 6304016 Free PMC article.
-
Endonuclease IV cleaves apurinic/apyrimidinic sites in single-stranded DNA and its application for biosensing.Analyst. 2016 Jul 21;141(14):4373-80. doi: 10.1039/c6an00738d. Epub 2016 May 17. Analyst. 2016. PMID: 27186607
Cited by
-
In vitro characterization of the site-specific recombination system based on actinophage TG1 integrase.Mol Genet Genomics. 2009 Dec;282(6):607-16. doi: 10.1007/s00438-009-0490-2. Epub 2009 Oct 16. Mol Genet Genomics. 2009. PMID: 19834741
-
A hexanucleotide sequence (dC1-dC6 tract) restricts the dC-specific cleavage of single-stranded DNA by endonuclease IV of bacteriophage T4.Nucleic Acids Res. 2007;35(19):6681-9. doi: 10.1093/nar/gkm721. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17940096 Free PMC article.
-
Treatment of E. coli Infections with T4-Related Bacteriophages Belonging to Class Caudoviricetes: Selecting Phage on the Basis of Their Generalized Transduction Capability.Viruses. 2025 May 14;17(5):701. doi: 10.3390/v17050701. Viruses. 2025. PMID: 40431712 Free PMC article.
References
-
- Wilson GG, Young KY, Edlin GJ, Konigsberg W. High-frequency generalized transduction by bacteriophage T4. Nature. 1979;280:80–82. - PubMed

