Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics
- PMID: 17913750
- PMCID: PMC2190718
- DOI: 10.1093/nar/gkm711
Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics
Abstract
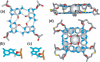
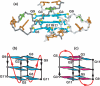
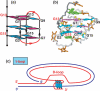
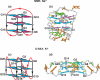
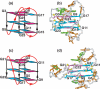
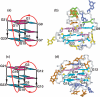
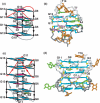
Guanine-rich DNA sequences can form G-quadruplexes stabilized by stacked G-G-G-G tetrads in monovalent cation-containing solution. The length and number of individual G-tracts and the length and sequence context of linker residues define the diverse topologies adopted by G-quadruplexes. The review highlights recent solution NMR-based G-quadruplex structures formed by the four-repeat human telomere in K(+) solution and the guanine-rich strands of c-myc, c-kit and variant bcl-2 oncogenic promoters, as well as a bimolecular G-quadruplex that targets HIV-1 integrase. Such structure determinations have helped to identify unanticipated scaffolds such as interlocked G-quadruplexes, as well as novel topologies represented by double-chain-reversal and V-shaped loops, triads, mixed tetrads, adenine-mediated pentads and hexads and snap-back G-tetrad alignments. The review also highlights the recent identification of guanine-rich sequences positioned adjacent to translation start sites in 5'-untranslated regions (5'-UTRs) of RNA oncogenic sequences. The activity of the enzyme telomerase, which maintains telomere length, can be negatively regulated through G-quadruplex formation at telomeric ends. The review evaluates progress related to ongoing efforts to identify small molecule drugs that bind and stabilize distinct G-quadruplex scaffolds associated with telomeric and oncogenic sequences, and outlines progress towards identifying recognition principles based on several X-ray-based structures of ligand-G-quadruplex complexes.
Figures







References
-
- Zimmerman SB, Cohen GH, Davies DR. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. - PubMed
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Wang Y, Patel DJ. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. - PubMed
-
- Laughlan G, Murchie AI, Norman DG, Moore MH, Moody PC, Lilley DM, Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

