Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein
- PMID: 17913801
- PMCID: PMC2168867
- DOI: 10.1128/JVI.00732-07
Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein
Abstract
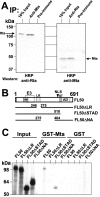
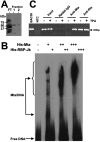
The Kaposi's sarcoma-associated herpesvirus (KSHV) Mta protein, encoded by open reading frame 57, is a transactivator of gene expression that is essential for productive viral replication. Previous studies have suggested both transcriptional and posttranscriptional roles for Mta, but little is known regarding Mta's transcriptional function. In this study, we demonstrate that Mta cooperates with the KSHV lytic switch protein, Rta, to reactivate KSHV from latency, but Mta has little effect on reactivation when expressed alone. We demonstrate that the Mta and Rta proteins are expressed with similar but distinct kinetics during KSHV reactivation. In single-cell analyses, Mta expression coincides tightly with progression to full viral reactivation. We demonstrate with promoter reporter assays that while Rta activates transcription in all cell lines tested, Mta's ability to transactivate promoters, either alone or synergistically with Rta, is cell and promoter specific. In particular, Mta robustly transactivates the nut-1/PAN promoter independently of Rta in 293 and Akata-31 cells. Using nuclear run-on assays, we demonstrate that Mta stimulates transcriptional initiation in 293 cells. Rta and Mta physically interact in infected cell extracts, and this interaction requires the intact leucine repeat and central region of Rta in vitro. We demonstrate that Mta also binds to the nut-1/PAN promoter DNA in vitro and in infected cells. An Mta mutant with a lesion in a putative A/T hook domain is altered in DNA binding and debilitated in transactivation. We propose that one molecular mechanism of Mta-mediated transactivation is a direct effect on transcription by direct and indirect promoter association.
Figures





Similar articles
-
KSHV reactivation and novel implications of protein isomerization on lytic switch control.Viruses. 2015 Jan 12;7(1):72-109. doi: 10.3390/v7010072. Viruses. 2015. PMID: 25588053 Free PMC article. Review.
-
CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters.J Virol. 2003 Sep;77(17):9590-612. doi: 10.1128/jvi.77.17.9590-9612.2003. J Virol. 2003. PMID: 12915572 Free PMC article.
-
Direct interactions of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta protein with the cellular protein octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation.J Virol. 2007 Aug;81(16):8451-67. doi: 10.1128/JVI.00265-07. Epub 2007 May 30. J Virol. 2007. PMID: 17537858 Free PMC article.
-
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential.J Virol. 2016 Dec 16;91(1):e01434-16. doi: 10.1128/JVI.01434-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795414 Free PMC article.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
Cited by
-
KSHV reactivation and novel implications of protein isomerization on lytic switch control.Viruses. 2015 Jan 12;7(1):72-109. doi: 10.3390/v7010072. Viruses. 2015. PMID: 25588053 Free PMC article. Review.
-
Identification of the physiological gene targets of the essential lytic replicative Kaposi's sarcoma-associated herpesvirus ORF57 protein.J Virol. 2015 Feb;89(3):1688-702. doi: 10.1128/JVI.02663-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410858 Free PMC article.
-
Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program.Virology. 2008 Sep 30;379(2):234-44. doi: 10.1016/j.virol.2008.06.043. Epub 2008 Aug 5. Virology. 2008. PMID: 18684478 Free PMC article.
-
New insights into the expression and functions of the Kaposi's sarcoma-associated herpesvirus long noncoding PAN RNA.Virus Res. 2016 Jan 2;212:53-63. doi: 10.1016/j.virusres.2015.06.012. Epub 2015 Jun 21. Virus Res. 2016. PMID: 26103097 Free PMC article. Review.
-
KSHV ORF57, a protein of many faces.Viruses. 2015 Feb 10;7(2):604-33. doi: 10.3390/v7020604. Viruses. 2015. PMID: 25674768 Free PMC article. Review.
References
-
- Agresti, A., and M. E. Bianchi. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170-178. - PubMed
-
- Albrecht, R. A., S. K. Kim, and D. J. O'Callaghan. 2005. The EICP27 protein of equine herpesvirus 1 is recruited to viral promoters by its interaction with the immediate-early protein. Virology 333:74-87. - PubMed
-
- Albrecht, R. A., S. K. Kim, Y. Zhang, Y. Zhao, and D. J. O'Callaghan. 2004. The equine herpesvirus 1 EICP27 protein enhances gene expression via an interaction with TATA box-binding protein. Virology 324:311-326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

