Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA
- PMID: 17913815
- PMCID: PMC2168881
- DOI: 10.1128/JVI.01244-07
Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA
Abstract
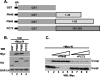
The nucleocapsid (N) protein of nonsegmented negative-strand (NNS) RNA viruses, when expressed in eukaryotic cells, aggregates and forms nucleocapsid-like complexes with cellular RNAs. The phosphoprotein (P) has been shown to prevent such aggregation by forming a soluble complex with the N protein free from cellular RNAs (designated N(0)). The N(0)-P complex presumably mediates specific encapsidation of the viral genome RNA. The precise mechanism by which the P protein carries out this function remains unclear. Here, by using a series of deleted and truncated mutant forms of the P protein of vesicular stomatitis virus (VSV), Indiana serotype, we present evidence that the N-terminal 11 to 30 amino acids (aa) of the P protein are essential in keeping the N protein soluble. Furthermore, glutathione S-transferase fused to the N-terminal 40 aa by itself is able to form the N(0)-P complex. Interestingly, the N-terminal 40-aa stretch failed to interact with the viral genome N-RNA template whereas the C-terminal 72 aa of the P protein interacted specifically with the latter. With an in vivo VSV minigenome transcription system, we further show that a deletion mutant form of P (PDelta1-10) lacking the N-terminal 10 aa which is capable of forming the N(0)-P complex was unable to support VSV minigenome transcription, although it efficiently supported transcription in vitro in a transcription-reconstitution reaction when used as purified protein. However, the same mutant protein complemented minigenome transcription when expressed together with a transcription-defective P deletion mutant protein containing N-terminal aa 1 to 210 (PDeltaII+III). Since the minigenome RNA needs to be encapsidated before transcription ensues, it seems that the entire N-terminal 210 aa are required for efficient genome RNA encapsidation. Taking these results together, we conclude that the N-terminal 11 to 30 aa are required for N(0)-P complex formation but the N-terminal 210 aa are required for genome RNA encapsidation.
Figures


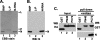

Similar articles
-
Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA.Virology. 1997 Nov 10;238(1):103-14. doi: 10.1006/viro.1997.8823. Virology. 1997. PMID: 9375014
-
Oligomerization of the Vesicular Stomatitis Virus Phosphoprotein Is Dispensable for mRNA Synthesis but Facilitates RNA Replication.J Virol. 2020 Jun 16;94(13):e00115-20. doi: 10.1128/JVI.00115-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32321813 Free PMC article.
-
Importance of hydrogen bond contacts between the N protein and RNA genome of vesicular stomatitis virus in encapsidation and RNA synthesis.J Virol. 2010 Feb;84(4):1741-51. doi: 10.1128/JVI.01803-09. Epub 2009 Dec 9. J Virol. 2010. PMID: 20007268 Free PMC article.
-
The Nucleocapsid of Paramyxoviruses: Structure and Function of an Encapsidated Template.Viruses. 2021 Dec 9;13(12):2465. doi: 10.3390/v13122465. Viruses. 2021. PMID: 34960734 Free PMC article. Review.
-
Reviewing Chandipura: a vesiculovirus in human epidemics.Biosci Rep. 2007 Oct;27(4-5):275-98. doi: 10.1007/s10540-007-9054-z. Biosci Rep. 2007. PMID: 17610154 Free PMC article. Review.
Cited by
-
Inclusion Body Fusion of Human Parainfluenza Virus Type 3 Regulated by Acetylated α-Tubulin Enhances Viral Replication.J Virol. 2017 Jan 18;91(3):e01802-16. doi: 10.1128/JVI.01802-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881643 Free PMC article.
-
Sumoylation of Human Parainfluenza Virus Type 3 Phosphoprotein Correlates with A Reduction in Viral Replication.Virol Sin. 2021 Jun;36(3):438-448. doi: 10.1007/s12250-020-00314-2. Epub 2020 Nov 16. Virol Sin. 2021. PMID: 33197004 Free PMC article.
-
RNA synthetic mechanisms employed by diverse families of RNA viruses.Wiley Interdiscip Rev RNA. 2013 Jul-Aug;4(4):351-67. doi: 10.1002/wrna.1164. Epub 2013 Apr 18. Wiley Interdiscip Rev RNA. 2013. PMID: 23606593 Free PMC article. Review.
-
Newly identified phosphorylation site in the vesicular stomatitis virus P protein is required for viral RNA synthesis.J Virol. 2014 Feb;88(3):1461-72. doi: 10.1128/JVI.02384-13. Epub 2013 Nov 20. J Virol. 2014. PMID: 24257610 Free PMC article.
-
Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication.J Virol. 2014 Apr;88(8):4414-22. doi: 10.1128/JVI.03673-13. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501400 Free PMC article.
References
-
- Albertini, A. A., A. K. Wernimont, T. Muziol, R. B. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360-363. - PubMed
-
- Basak, S., T. Raha, D. Chattopadhyay, A. Majumder, M. S. Shaila, and D. J. Chattopadhyay. 2003. Leader RNA binding ability of Chandipura virus P protein is regulated by its phosphorylation status: a possible role in genome transcription-replication switch. Virology 307:372-385. - PubMed
-
- Bhattacharya, R., S. Basak, and D. J. Chattopadhyay. 2006. Initiation of encapsidation as evidenced by deoxycholate-treated nucleocapsid protein in the Chandipura virus life cycle. Virology 349:197-211. - PubMed
-
- Bhella, D., A. Ralph, L. B. Murphy, and R. P. Yeo. 2002. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 83:1831-1839. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

